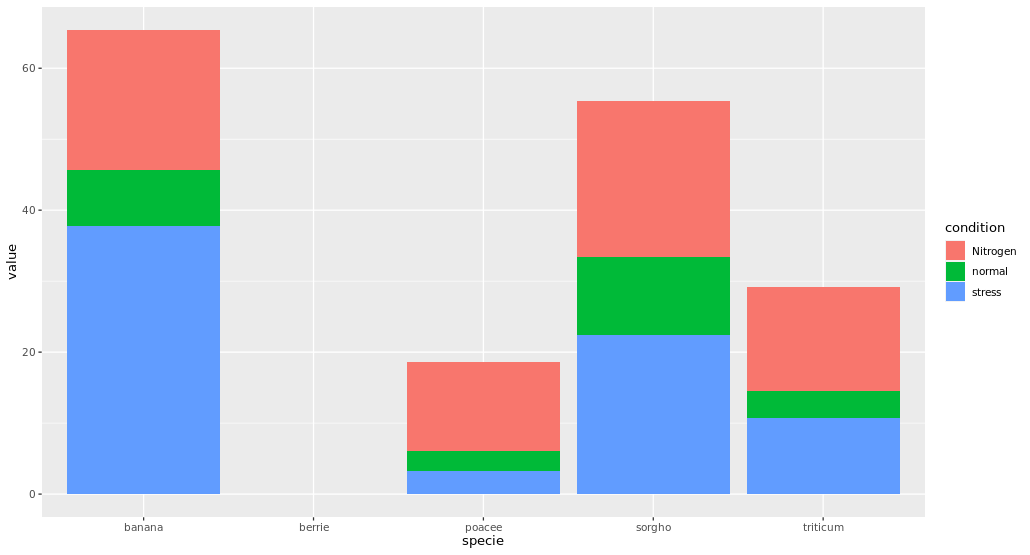
The below code generates a table however, when I plot the table the resulting figure y-axis does not match the values present in the data frame.
set.seed(1)
specie <- c(rep("sorgho" , 3) , rep("poacee" , 3) , rep("banana" , 3) , rep("triticum" , 3), rep('berrie', 3) )
condition <- rep(c("normal" , "stress" , "Nitrogen") , 5)
value <- append( abs(rnorm(12 , 0 , 15)), rep(NA,3))
data <- data.frame(specie,condition,value)
> data
specie condition value
1 sorgho normal 9.396807
2 sorgho stress 2.754650
3 sorgho Nitrogen 12.534429
4 poacee normal 23.929212
5 poacee stress 4.942617
6 poacee Nitrogen 12.307026
7 banana normal 7.311436
8 banana stress 11.074871
9 banana Nitrogen 8.636720
10 triticum normal 4.580826
11 triticum stress 22.676718
12 triticum Nitrogen 5.847649
13 berrie normal NA
14 berrie stress NA
15 berrie Nitrogen NA
ggplot(data, aes(fill=condition, y=value, x=specie))
geom_bar(position="stack", stat="identity")
CodePudding user response:
when I run your code, as above, I get proper results, see below. Maybe try a new R session
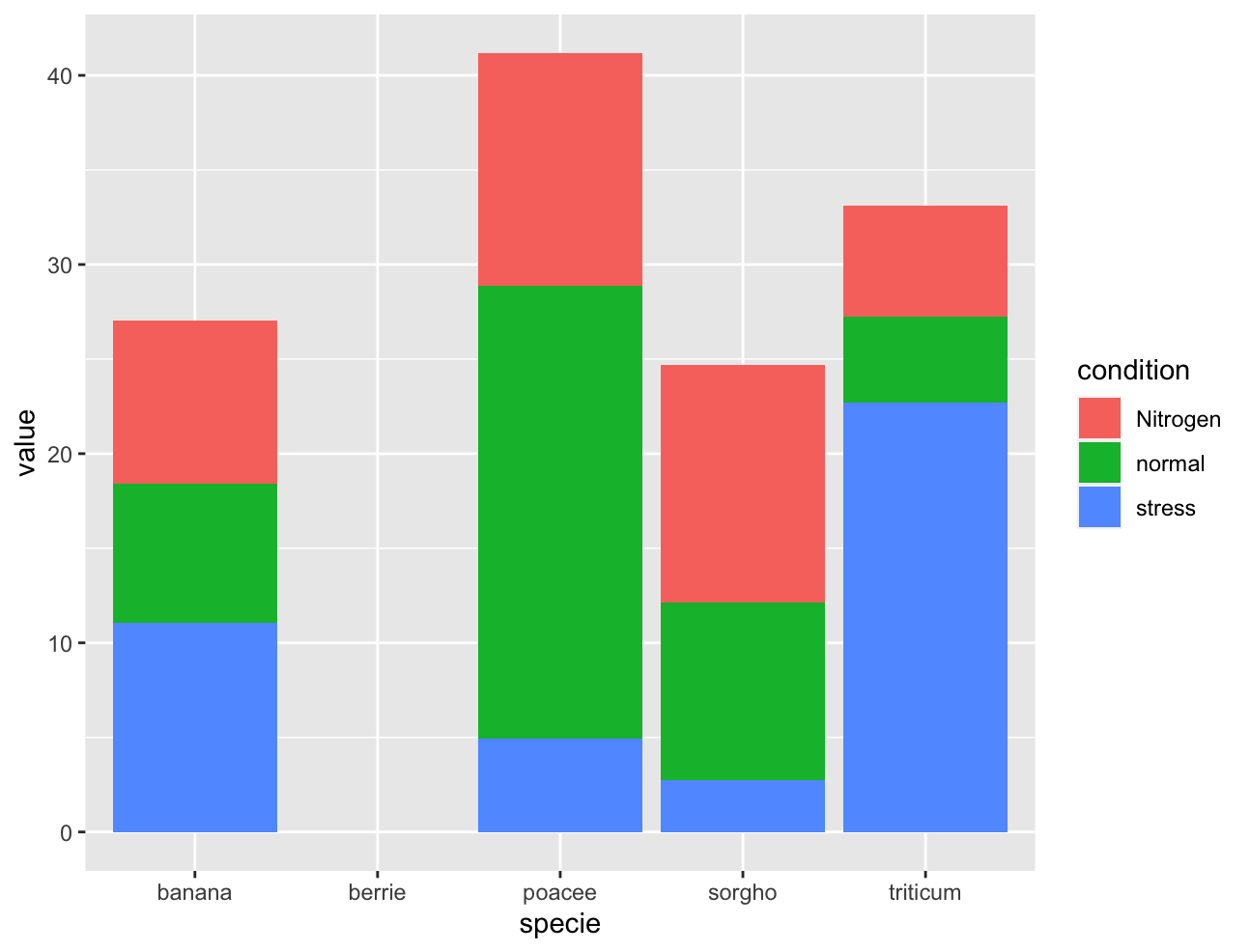
My table:
> data
specie condition value
1 sorgho normal 9.396807
2 sorgho stress 2.754650
3 sorgho Nitrogen 12.534429
4 poacee normal 23.929212
5 poacee stress 4.942617
6 poacee Nitrogen 12.307026
7 banana normal 7.311436
8 banana stress 11.074871
9 banana Nitrogen 8.636720
10 triticum normal 4.580826
11 triticum stress 22.676718
12 triticum Nitrogen 5.847649
13 berrie normal NA
14 berrie stress NA
15 berrie Nitrogen NA