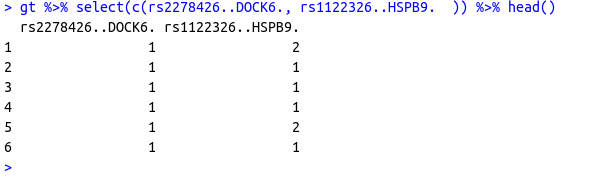
I have a R dataframe with values of certain columns (SNPs - Single Nucleotide Polymorphism ) are encoded as 1,2 and 3 like the following.. (just selected 2 columns out of 100s)
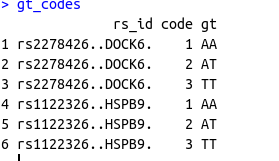
I have another data frame for the genotypes of these codes for each SNP as the following
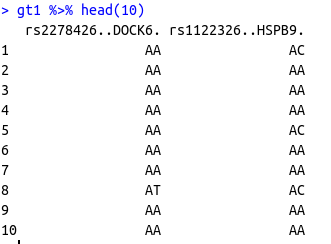
Now I want to decode the codes 1,2 and 3 in the dataframe gt according to the dataframe gt_codes and make a data frame like the following
I have given sample data as dput. Can someone please help me.
structure(list(rs2278426..DOCK6. = c(1L, 1L, 1L, 1L, 1L, 1L,
1L, 2L, 1L, 1L), rs1122326..HSPB9. = c(2L, 1L, 1L, 1L, 2L, 1L,
1L, 2L, 1L, 1L)), row.names = c(NA, 10L), class = "data.frame")
structure(list(rs_id = c("rs2278426..DOCK6.", "rs2278426..DOCK6.",
"rs2278426..DOCK6.", "rs1122326..HSPB9.", "rs1122326..HSPB9.",
"rs1122326..HSPB9."), code = c(1, 2, 3, 1, 2, 3), gt = c("AA",
"AT", "TT", "AA", "AT", "TT")), class = "data.frame", row.names = c(NA,
-6L))
CodePudding user response:
Here is a solution based on
- reshaping your original data from wide to long,
- doing a join with the mapping table
gt_codesto replace old with new values, and then - reshaping data again back from long to wide.
library(dplyr)
library(tidyr)
df %>%
mutate(row = 1:n()) %>%
pivot_longer(-row, names_to = "rs_id", values_to = "code") %>%
left_join(gt_codes, by = c("rs_id", "code")) %>%
select(-code) %>%
pivot_wider(names_from = "rs_id", values_from = "gt") %>%
select(-row)
## A tibble: 10 × 2
# rs2278426..DOCK6. rs1122326..HSPB9.
# <chr> <chr>
# 1 AA AC
# 2 AA AA
# 3 AA AA
# 4 AA AA
# 5 AA AC
# 6 AA AA
# 7 AA AA
# 8 AT AC
# 9 AA AA
#10 AA AA
Sample data
gt_codes <- data.frame(
rs_id = c(rep("rs2278426..DOCK6.", 3), rep("rs1122326..HSPB9.", 3)),
code = c(1:3, 1:3),
gt = c("AA", "AT", "TT", "AA", "AC", "CC"))
df <- structure(list(rs2278426..DOCK6. = c(1L, 1L, 1L, 1L, 1L, 1L,
1L, 2L, 1L, 1L), rs1122326..HSPB9. = c(2L, 1L, 1L, 1L, 2L, 1L,
1L, 2L, 1L, 1L)), row.names = c(NA, 10L), class = "data.frame")