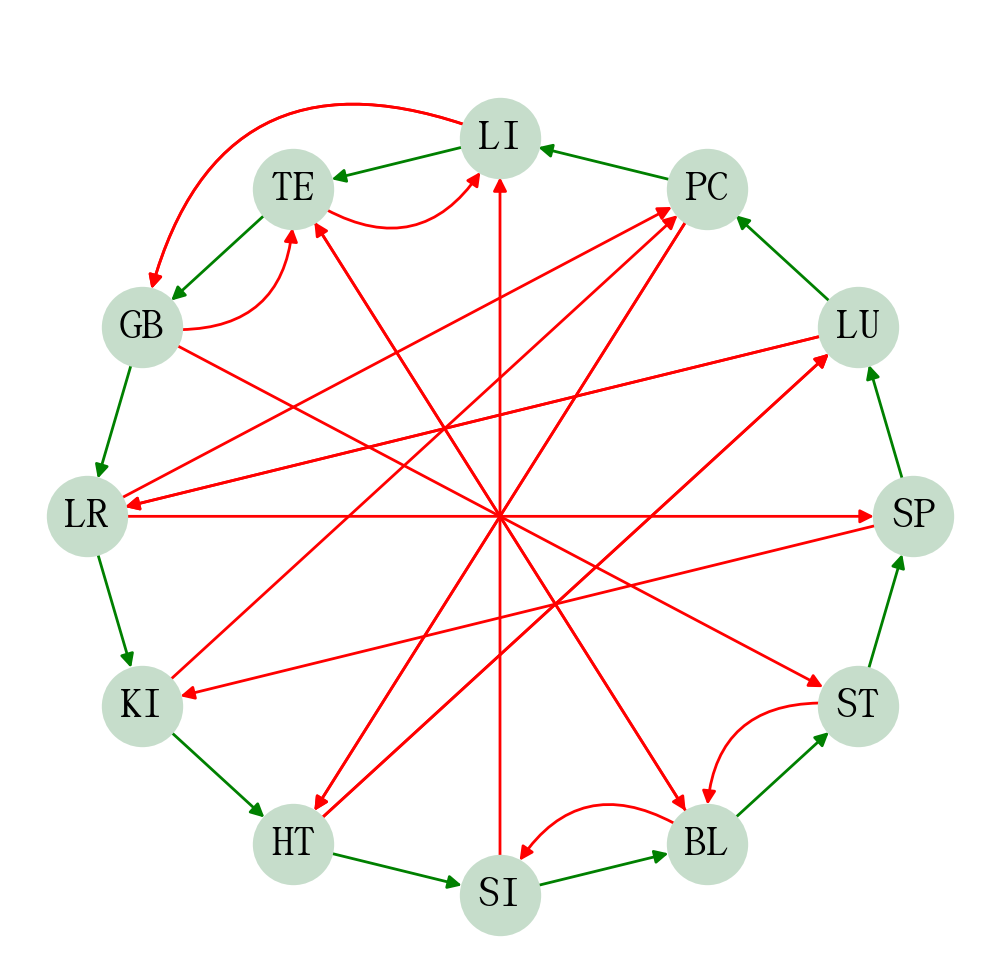
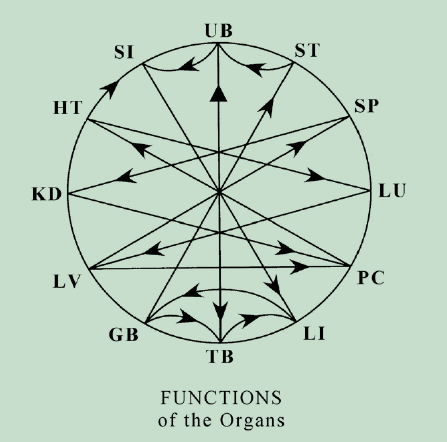
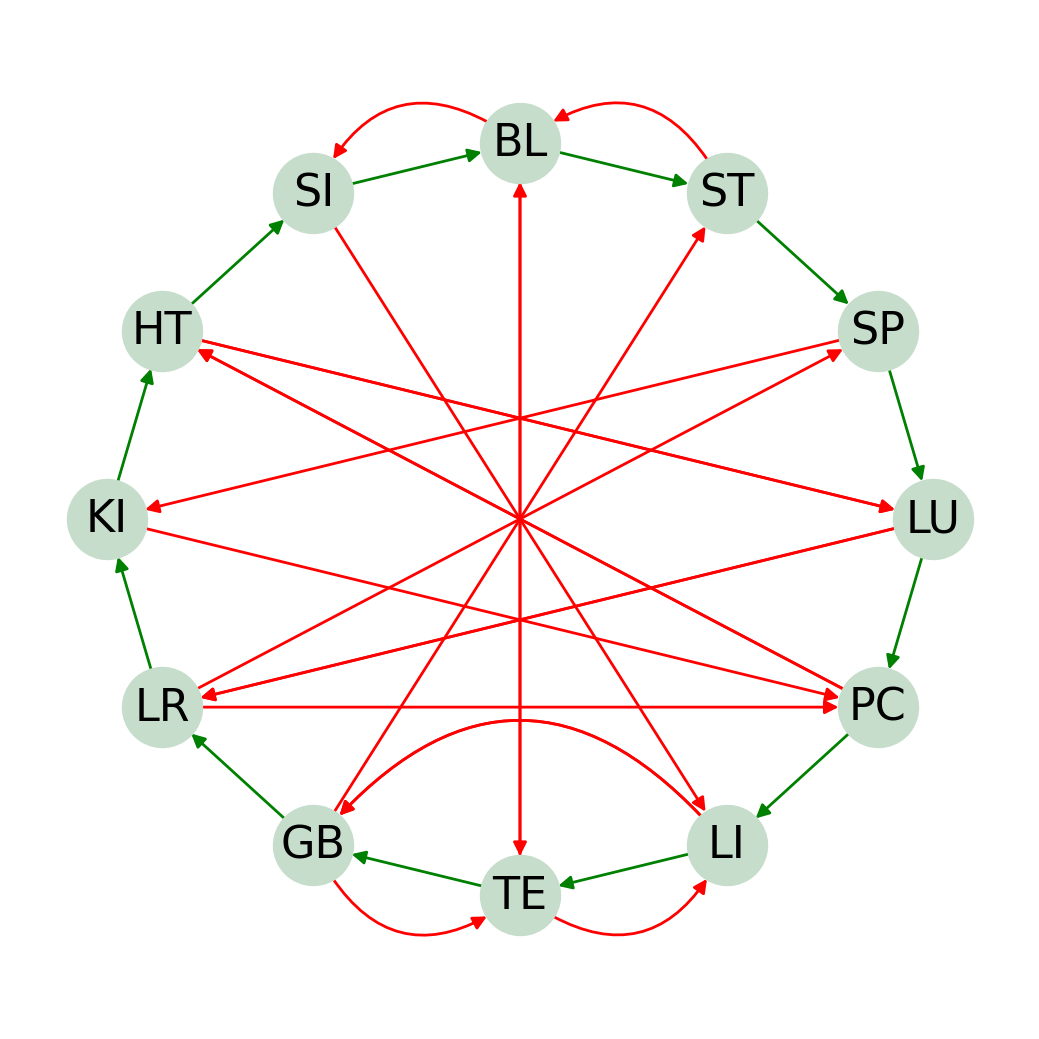
The following code generates a circular directed graph with networkx.
from matplotlib import pyplot as plt
import networkx as nx
def make_cyclic_edge(lst):
cyclic = []
for i, elem in enumerate(lst):
if i 1 < len(lst):
cyclic.append((elem, lst[i 1]))
else:
cyclic.append((elem, lst[0]))
return cyclic
def cycle_diagram(generate, inhibit, organ_func=False, plot=False):
"""Generate element cycle diagram with Networkx. """
G = nx.MultiDiGraph()
for pair in generate:
G.add_edge(*pair, color="g", group="generate")
for pair in inhibit:
G.add_edge(*pair, color="r", group="inhibit")
pos = nx.circular_layout(G, center=(0, 0))
edges_to_adjust = []
if organ_func:
fig = plt.figure(2, figsize=(5, 5), dpi=200)
edges_to_adjust = [("LI", "GB", 0), ("LI", "GB", 1), ("GB", "TE", 0), ("TE", "LI", 0),
("ST", "BL", 0), ("BL", "SI", 0)]
for edge in edges_to_adjust:
G.remove_edge(*edge)
color = list(nx.get_edge_attributes(G, 'color').values())
if organ_func:
nx.draw(
G, pos=pos,
with_labels=True,
font_size=16,
node_size=800,
node_color='#C6DDCB',
connectionstyle=f"arc3, rad=0.0",
edge_color=color,
)
nx.draw_networkx_edges(G,
pos=pos,
node_size=800,
edgelist=edges_to_adjust,
connectionstyle=f"arc3, rad=0.6",
edge_color=["r"]*len(edges_to_adjust),
)
else:
fig = plt.figure(1, figsize=(4, 4), dpi=200)
nx.draw(
G, pos=pos,
with_labels=True,
font_family='AR PL KaitiM Big5',
font_size=16,
node_size=800,
node_color='#C6DDCB',
connectionstyle=f"arc3, rad=0.235",
edge_color=color,
)
if plot:
plt.show()
return G
ORGAN_FUNC = (
"SP",
"LU",
"PC",
"LI",
"TE",
"GB",
"LR",
"KI",
"HT",
"SI",
"BL",
"ST",
)
ORGAN_FUNC_GEN = make_cyclic_edge(ORGAN_FUNC)
ORGAN_FUNC_INHIBIT_A = (
"BL",
"SI",
"LI",
"GB",
"ST",
)
ORGAN_FUNC_INHIBIT_Am = (
"LI",
"GB",
"TE",
)
ORGAN_FUNC_INHIBIT_B = (
"SP",
"KI",
"PC",
"HT",
"LU",
"LR",
)
ORGAN_FUNC_INHIBIT_C = (
"LR",
"PC",
"HT",
"LU",
)
ORGAN_FUNC_INHIBIT_D = (
"BL",
"TE",
)
ORGAN_FUNC_inhibit_list = [
ORGAN_FUNC_INHIBIT_A,
ORGAN_FUNC_INHIBIT_Am,
ORGAN_FUNC_INHIBIT_B,
ORGAN_FUNC_INHIBIT_C,
ORGAN_FUNC_INHIBIT_D,
]
ORGAN_FUNC_INHIBIT = []
for cycle in ORGAN_FUNC_inhibit_list:
ORGAN_FUNC_INHIBIT = make_cyclic_edge(cycle)
ORGAN_FUNC_CYCLE = cycle_diagram(ORGAN_FUNC_GEN, ORGAN_FUNC_INHIBIT, organ_func=True, plot=True)
I would like to:
- Make the arrows along its circumference run clockwise.
- Rotate the graph so that
"BL"is at the top.
like so (organ code-names differ slightly):
CodePudding user response:
It's not the most elegant, but here's one approach that works. I made the following changes to your cycle_diagram function. I added optional arguments top (to specify the top node) and flip (to specify whether to flip coordinates horizontally). Within the code, I added the following in after the layout pos is defined.
if top:
c,s = pos[top]
R_mat = np.array([[s,-c],[c,s]])
for k,v in pos.items():
pos[k] = R_mat@v
if flip:
for k,v in pos.items():
pos[k][0] *= -1
R_mat is a rotation matrix that rotates the point at pos[top] to the point (0,1), assuming that all points in the layout have magnitude 1 (which is indeed the case when the circular_layout function is used).
Here's the full script, with the additions.
from matplotlib import pyplot as plt
import networkx as nx
import numpy as np
def make_cyclic_edge(lst):
cyclic = []
for i, elem in enumerate(lst):
if i 1 < len(lst):
cyclic.append((elem, lst[i 1]))
else:
cyclic.append((elem, lst[0]))
return cyclic
def cycle_diagram(generate, inhibit, organ_func=False, plot=False, top=None, flip=False):
"""Generate element cycle diagram with Networkx. """
G = nx.MultiDiGraph()
for pair in generate:
G.add_edge(*pair, color="g", group="generate")
for pair in inhibit:
G.add_edge(*pair, color="r", group="inhibit")
pos = nx.circular_layout(G, center=(0, 0))
if top:
c,s = pos[top]
R_mat = np.array([[s,-c],[c,s]])
for k,v in pos.items():
pos[k] = R_mat@v
if flip:
for k,v in pos.items():
pos[k][0] *= -1
edges_to_adjust = []
if organ_func:
fig = plt.figure(2, figsize=(5, 5), dpi=200)
edges_to_adjust = [("LI", "GB", 0), ("LI", "GB", 1), ("GB", "TE", 0), ("TE", "LI", 0),
("ST", "BL", 0), ("BL", "SI", 0)]
for edge in edges_to_adjust:
G.remove_edge(*edge)
color = list(nx.get_edge_attributes(G, 'color').values())
if organ_func:
nx.draw(
G, pos=pos,
with_labels=True,
font_size=16,
node_size=800,
node_color='#C6DDCB',
connectionstyle=f"arc3, rad=0.0",
edge_color=color,
)
nx.draw_networkx_edges(G,
pos=pos,
node_size=800,
edgelist=edges_to_adjust,
connectionstyle=f"arc3, rad=0.6",
edge_color=["r"]*len(edges_to_adjust),
)
else:
fig = plt.figure(1, figsize=(4, 4), dpi=200)
nx.draw(
G, pos=pos,
with_labels=True,
font_family='AR PL KaitiM Big5',
font_size=16,
node_size=800,
node_color='#C6DDCB',
connectionstyle=f"arc3, rad=0.235",
edge_color=color,
)
if plot:
plt.show()
return G
ORGAN_FUNC = (
"SP",
"LU",
"PC",
"LI",
"TE",
"GB",
"LR",
"KI",
"HT",
"SI",
"BL",
"ST",
)
ORGAN_FUNC_GEN = make_cyclic_edge(ORGAN_FUNC)
ORGAN_FUNC_INHIBIT_A = (
"BL",
"SI",
"LI",
"GB",
"ST",
)
ORGAN_FUNC_INHIBIT_Am = (
"LI",
"GB",
"TE",
)
ORGAN_FUNC_INHIBIT_B = (
"SP",
"KI",
"PC",
"HT",
"LU",
"LR",
)
ORGAN_FUNC_INHIBIT_C = (
"LR",
"PC",
"HT",
"LU",
)
ORGAN_FUNC_INHIBIT_D = (
"BL",
"TE",
)
ORGAN_FUNC_inhibit_list = [
ORGAN_FUNC_INHIBIT_A,
ORGAN_FUNC_INHIBIT_Am,
ORGAN_FUNC_INHIBIT_B,
ORGAN_FUNC_INHIBIT_C,
ORGAN_FUNC_INHIBIT_D,
]
ORGAN_FUNC_INHIBIT = []
for cycle in ORGAN_FUNC_inhibit_list:
ORGAN_FUNC_INHIBIT = make_cyclic_edge(cycle)
ORGAN_FUNC_CYCLE = cycle_diagram(ORGAN_FUNC_GEN, ORGAN_FUNC_INHIBIT,
organ_func=True, plot=True, top = "BL", flip = True)
The result I get: