Context
I have a dataset that has some NA in a variable that is being used for grouping (I know, shocking!). When I throw this data at my ggplot2 blender, these NA values are treated (by default) as a group/level, which is not what I want for this plot.
I am using a standardized layer configuration for all plots, so I (a) do all data wrangling once, for all plots that will be created and (b) configure all layers that are common to all plots. I save these standard setup in a "blank" gg object and then just call it for every plot with the specific variables required for each occasion.
So, if I remove these NA from the overall gg object, I will also loose valid observations for other plots, and unfortunately this would make the client unhappy (and I don't want that). Surely, I could create a separate data frame just for this plot, and everything would be nice and dandy, the client would be happy, everyone would be happy. This is exactly how I approached it.
Now that I have time to think, I am curious on what would be the smart ggplot2 way.
Reproducible example: the issue
gg <- airquality %>%
# create some NA values in the grouping var
slice_head(n = 150) %>%
mutate(group = as.character(sample(c(1:2, NA), 150, replace = TRUE))) %>%
# standard configs for multiple plots
ggplot()
theme_bw()
scale_fill_brewer(palette = "Paired")
One particular plot is giving me trouble
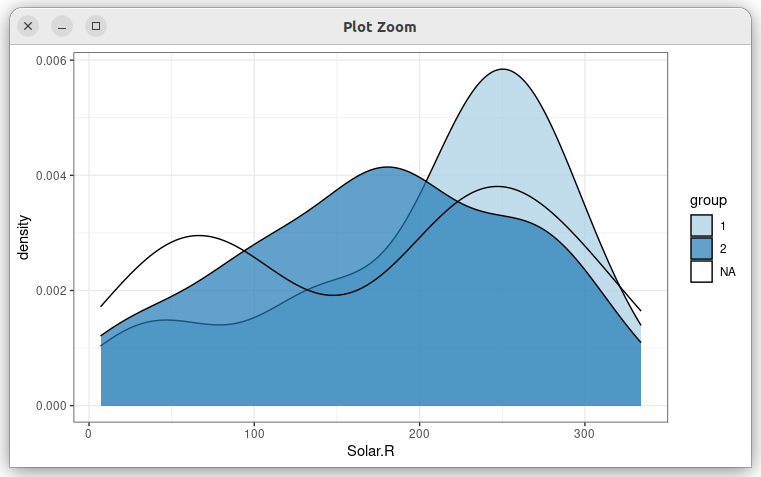
gg geom_density(aes(Solar.R, fill = group), alpha = .7)
Which results in:
What I have tried
I tried adding na.translate = FALSE in the scale layer as described in the docs. It helps, since it removes the NA group from the legend, but density curve is still there:
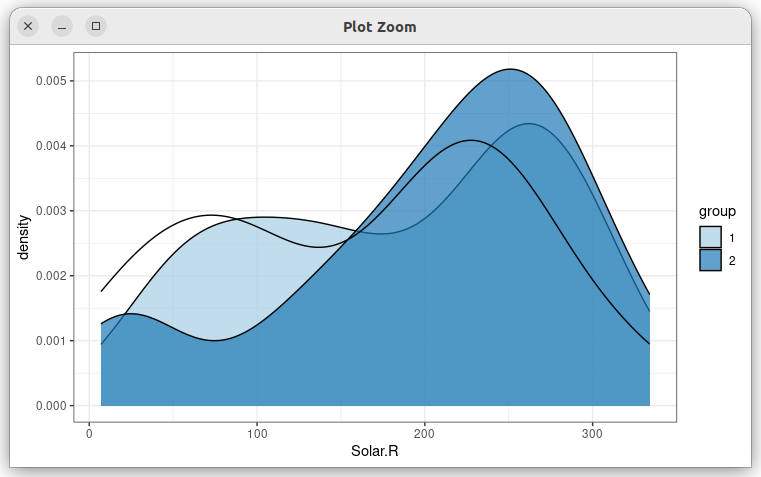
gg <- airquality %>%
# similar to above, except the scale
scale_fill_brewer(palette = "Paired", na.translate = FALSE)
gg geom_density(aes(Solar.R, fill = group), alpha = .7)
I also tried adding na.value = NA to the scale_fill function, but it didn't seem to make a difference. Also values like NULL and "blank" are not permitted by this particular scale.
Finally, I have seen geom_bar() examples with na.rm = TRUE in the aesthetic definition, but geom_density() refuses to accept this solution.
Question
- How do I drop this
NAlevel altogether from one particular plot that is sharing a standard configuration from other sibling plots, without disturbing the others?
I'm guessing there is some clever way of defining the aesthetics, or maybe another argument to scale_fill_*() that I'm not aware of.
CodePudding user response:
Perhaps one of these approaches would suit?
library(tidyverse)
set.seed(123)
test_data <- airquality %>%
# create some NA values in the grouping var
slice_head(n = 150) %>%
mutate(group = as.character(sample(c(1:2, NA), 150, replace = TRUE)))
# standard configs for multiple plots
gg <- ggplot(test_data)
theme_bw()
scale_fill_brewer(palette = "Paired")
# with NAs
gg geom_density(aes(Solar.R, fill = group), alpha = .7)
#> Warning: Removed 7 rows containing non-finite values (`stat_density()`).
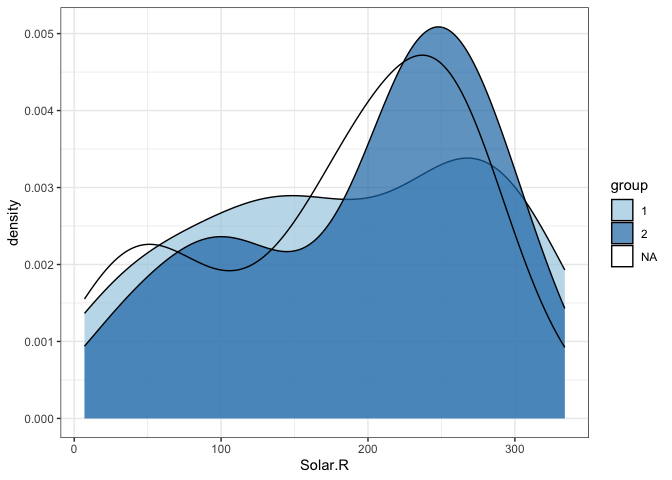
# without NAs
gg geom_density(data = test_data %>% filter(!is.na(group)),
aes(Solar.R, fill = group), alpha = .7)
#> Warning: Removed 5 rows containing non-finite values (`stat_density()`).
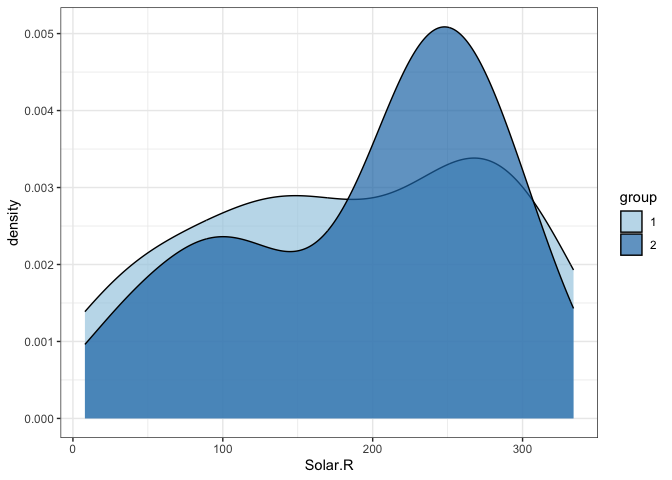
Or perhaps this method?
set.seed(123)
gg <- airquality %>%
# create some NA values in the grouping var
slice_head(n = 150) %>%
mutate(group = as.character(sample(c(1:2, NA), 150, replace = TRUE))) %>%
# standard configs for multiple plots
{ggplot(data = .)
theme_bw()
scale_fill_brewer(palette = "Paired")}
gg
geom_density(data = . %>% filter(!is.na(group)), aes(Solar.R, fill = group), alpha = .7)
#> Warning: Removed 5 rows containing non-finite values (`stat_density()`).
Created on 2022-12-06 with reprex v2.0.2