I am trying to visualize 3 categorical variables: a binary categorical variable between the treatment and control groups and before&treatmet. The main variable measures whether someone's views are sociotropic or egocentric, and below is my attempt in doing so using bar graphs, but I am open to other graphs to visualize the same variables.
data example:
print data example
dput(sample_n(socio_egotropic_graph, size = 5))
structure(list(date = structure(c(1346112000, 1335139200, 1318118400, 1349913600,
1339891200), tzone = "UTC", class = c("POSIXct", "POSIXt")),
sentiment_human_coded = c("negative",
"negative", "neutral", "negative", "negative"), economic_demand_complaint = c(1,
0, 0, 0, 0), collective_action = c(0, 0, 1, 0, 0), directed_to_whom = c("Private employer",
"N/A", NA, "N/A", "Private employer"), socio_egotropic = c("sociotropic",
"egocentric", "sociotropic", "egocentric", "egocentric"),
gender = c("female", "female", NA, "male", "female"), treatment_announcement = c("post",
"post", NA, "post", "post"), treatment_details = c("post",
"post", "pre", "post", "post"), treatment_implementation = c("pre",
"pre", "pre", "post", "pre"), month_year = structure(c(2012.58333333333,
2012.25, 2011.75, 2012.75, 2012.41666666667), class = "yearmon"),
group = c("treatment", "treatment", "control", "treatment",
"treatment")), row.names = c(NA, -5L), class = c("tbl_df",
"tbl", "data.frame"))
graph code:
socio_egotropic_graph |>
drop_na() |>
filter(socio_egotropic != "N/A") |>
select(socio_egotropic, treatment_details, group) |> # we're only interested in socio_egotropic
group_by(socio_egotropic) %>% # group data and
add_count(treatment_details) |> # add count of treatment_details
unique() |> # remove duplicates
ungroup() |> # remove grouping
group_by(treatment_details) |> # group by treatment_details
mutate(socio_egotropic_percentage = n/sum(n)) |> # ...calculating percentage
mutate(socio_egotropic = as.factor(socio_egotropic)) |> # change to factors so that ggplot treats...
mutate(am = as.factor(treatment_details)) |>
ggplot(aes(x = treatment_details, fill = socio_egotropic, y = socio_egotropic_percentage))
geom_bar(stat = "identity", position=position_dodge())
#scale_fill_grey()
xlab("Socio vs. egocentric emotions...")
ylab("Socio egocentric share")
theme(text=element_text(size=10))
scale_y_continuous(labels = percent_format(accuracy = 1))
theme(plot.title = element_text(size = 10, face = "bold"))
scale_x_discrete(limits = c("pre", "post"))
theme_bw()
Here is the output, while the code works, I am unable to show variation depending on treatment status, which is measured using the "group" variable.
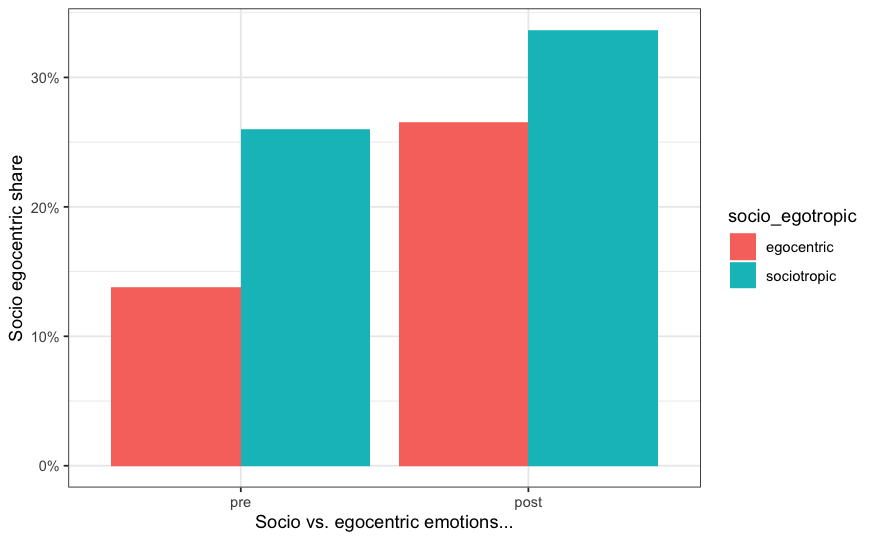
CodePudding user response:
One approach:
Since the dput output failed on my side, first construct an example dataset:
n = 40 ## sample data size
get_dummy <- function(choices, n = 40) sample(choices, n, TRUE)
set.seed(4711)
df <- data.frame(month_year = as.Date('2022-03-15') sample(-2:2, n, TRUE)
30 * sample(0:1, n, TRUE),
socio_egotropic = get_dummy(c('sociotropic', 'egocentric')),
treatment_details = get_dummy(c('pre', 'post')),
group = get_dummy(c('treatment', 'control'))
)
summarize data as frequency table (retain 'egocentric' trait only):
df_stats_egocentric <- df |>
## uncomment, if control values cannot be assigned to pre/post:
## mutate(treatment_details = ifelse(group == 'control',
## 'post', treatment_details)
## ) |>
count(socio_egotropic, treatment_details, group) |>
group_by(group, treatment_details) |>
mutate(prop = prop.table(n)) |>
filter(socio_egotropic == 'egocentric')
bullet plot, if control observations can also be assigned to pre- and post-treatment periods:
df_stats_egocentric |>
ggplot()
geom_col(data = . %>% filter(group == 'treatment'),
aes(treatment_details, prop),
alpha = .5)
geom_col(data = . %>% filter(group == 'control'),
aes(treatment_details, prop),
width = .01)
If control observations apply to both pre- and post-treatment effects, draw a horizontal reference line:
df_stats_egocentric |>
ggplot()
geom_col(data = . %>% filter(group == 'treatment'),
aes(treatment_details, prop),
alpha = .5)
geom_hline(data = . %>% filter(group == 'control'),
aes(yintercept = prop)
)
