I am running a mixed effects model on my dataset ,
library(lme4)
data(cake)
each dataset is a subset of a larger datsaet
subset(cake, recipe=="A")
subset(cake, recipe=="B")
subset(cake, recipe=="C")
I am using dlply to run my mixed effects model on each subset
MxM1 <- plyr::dlply(cake,
"recipe",
function(x)
lmer(angle ~ 1 (1|replicate) temperature,
data=x))
This gives me a list of summaries based on each subset of data.
I know how to display the summaries one at a time using gt_summary package
lm_cake$A %>%
tbl_regression() %>%
modify_column_hide(columns = ci) %>%
modify_column_unhide(columns = std.error)
lm_cake$B %>%
tbl_regression() %>%
modify_column_hide(columns = ci) %>%
modify_column_unhide(columns = std.error)
lm_cake$B %>%
tbl_regression() %>%
modify_column_hide(columns = ci) %>%
modify_column_unhide(columns = std.error)
I am not sure how to combine the results from all 3 objects (lm_cake$A, lm_cake$B, lm_cake$C) to display them as one summary table.
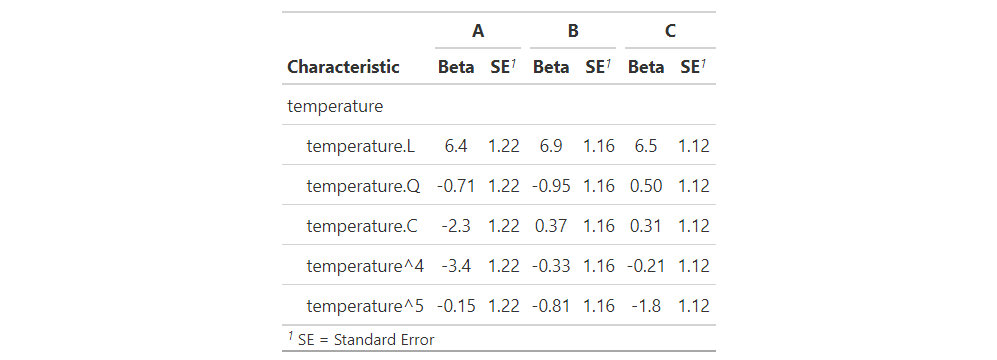
Model: A Model: B Model: C
Temperature Beta SE Beta SE Beta SE
Temperature. L
Temperature. Q
Temperature. C
Temperature^4
Temperature^5
Any suggestions or help is much apricated. Thanks.
CodePudding user response:
You need something like this?:
library(lme4)
data(cake)
library(dplyr)
library(broom)
library(broom.mixed)
cake %>%
mutate(recipe = as_factor(recipe)) %>%
group_by(recipe) %>%
group_split() %>%
map_dfr(.f = function(df){
lmer(angle ~ 1 (1|replicate) temperature,
data=df) %>%
tidy() %>%
add_column(recipe = unique(df$recipe), .before = 1)
})
A tibble: 24 × 7
recipe effect group term estimate std.error statistic
<fct> <chr> <chr> <chr> <dbl> <dbl> <dbl>
1 A fixed NA (Intercept) 33.1 1.42 23.3
2 A fixed NA temperature.L 6.43 1.22 5.26
3 A fixed NA temperature.Q -0.713 1.22 -0.583
4 A fixed NA temperature.C -2.33 1.22 -1.90
5 A fixed NA temperature^4 -3.35 1.22 -2.74
6 A fixed NA temperature^5 -0.151 1.22 -0.124
7 A ran_pars replicate sd__(Intercept) 5.16 NA NA
8 A ran_pars Residual sd__Observation 4.73 NA NA
9 B fixed NA (Intercept) 31.6 1.81 17.5
10 B fixed NA temperature.L 6.88 1.16 5.93
# … with 14 more rows
# ℹ Use `print(n = ...)` to see more rows
CodePudding user response:
You can also merge two or more gtsummary tables using the gtsummary::tbl_merge() function. Example below!
library(gtsummary)
#> #StandWithUkraine
library(lme4)
#> Loading required package: Matrix
data(cake)
MxM1 <-
plyr::dlply(
cake,
"recipe",
function(x) {
lmer(angle ~ 1 (1|replicate) temperature, data=x) %>%
tbl_regression() %>%
modify_column_hide(columns = ci) %>%
modify_column_unhide(columns = std.error)
}
)
# Merge all model summaries together with `tbl_merge()`
tbl <-
MxM1 %>%
tbl_merge(
tab_spanner = c("**A**", "**B**", "**C**")
)
 Created on 2022-12-17 with reprex v2.0.2
Created on 2022-12-17 with reprex v2.0.2
