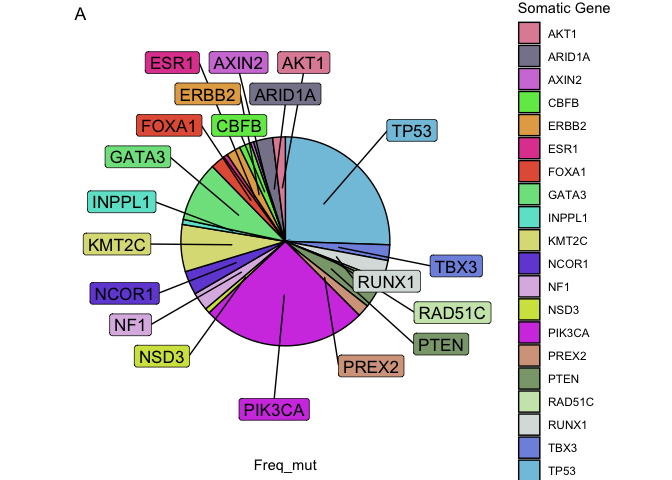
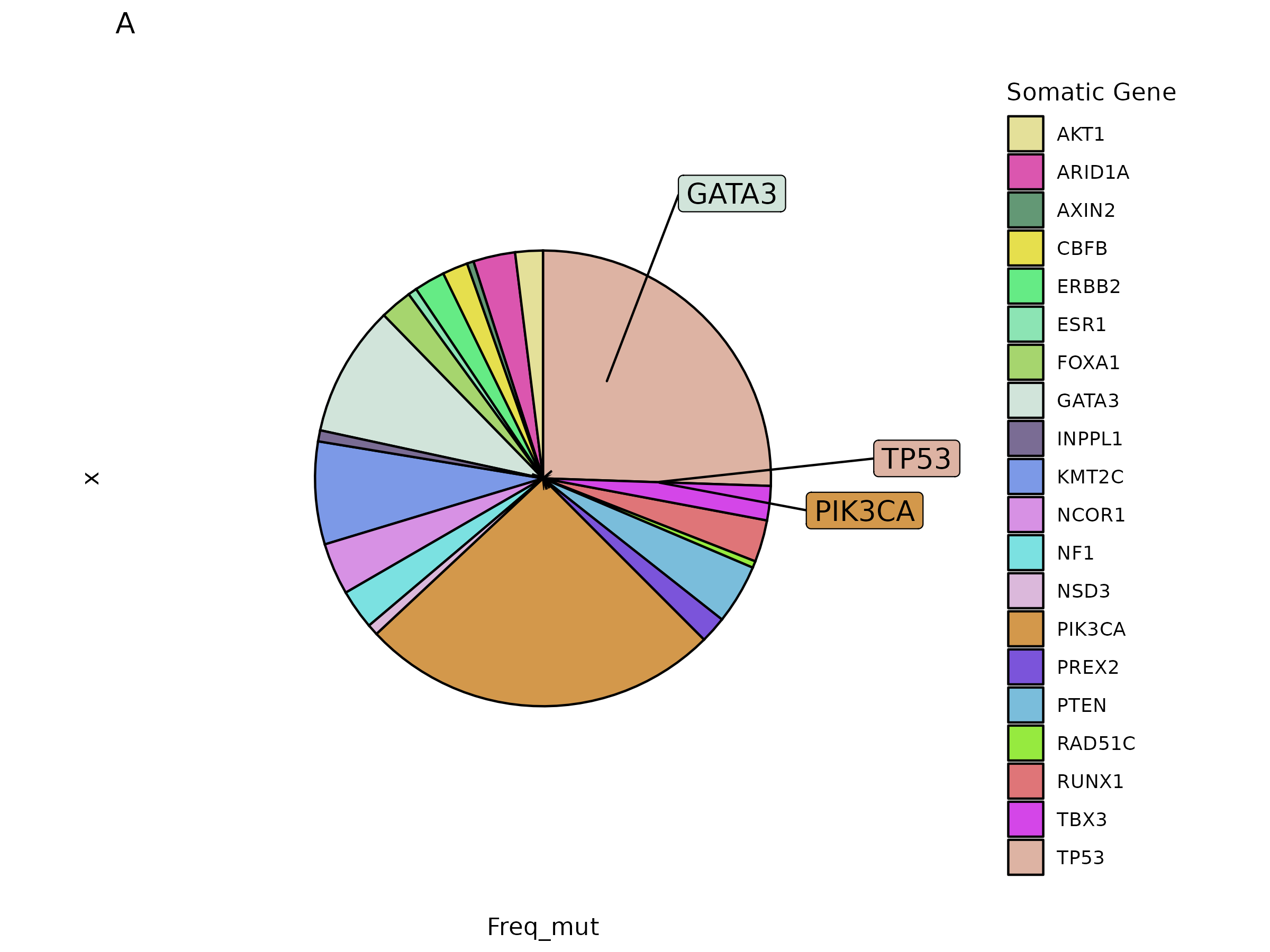
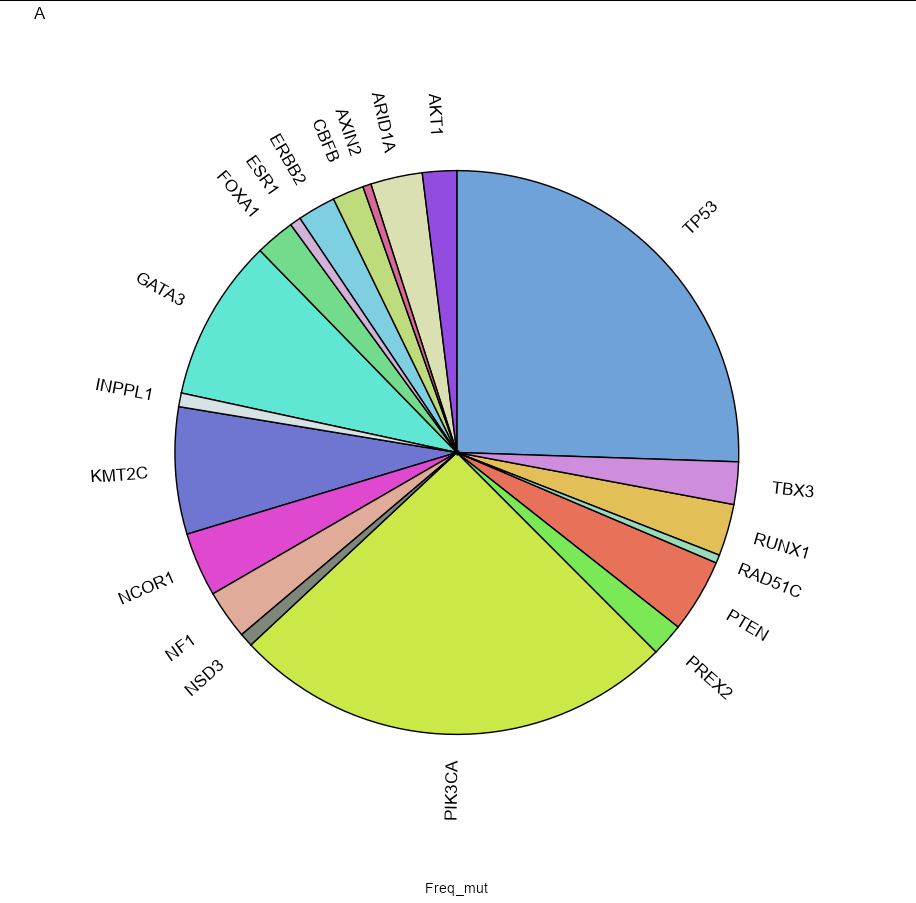
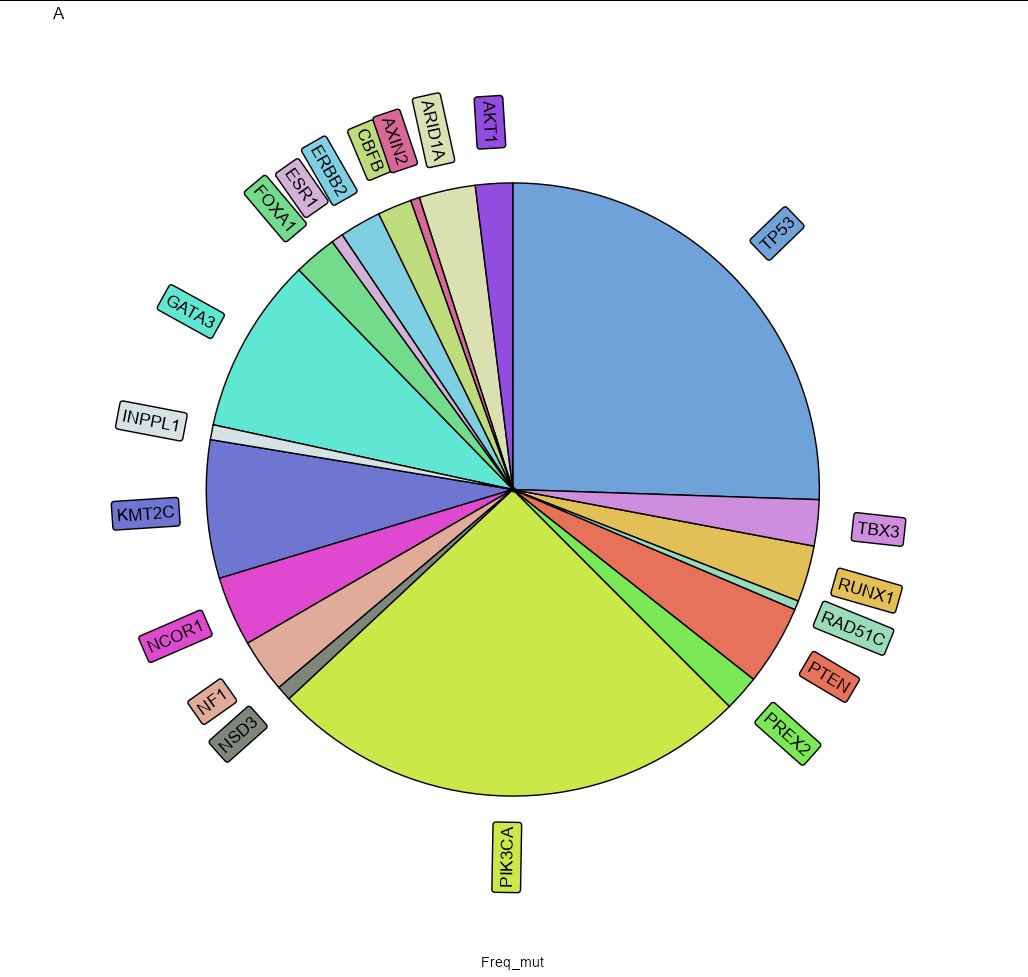
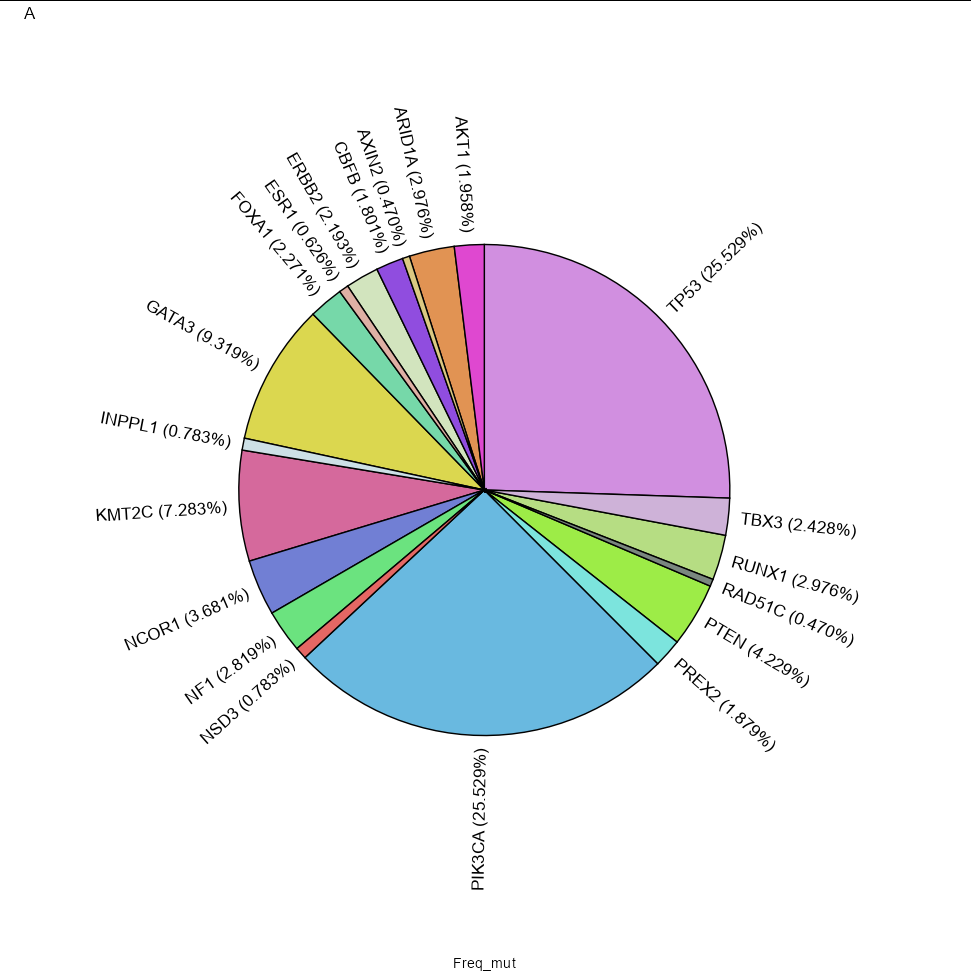
I am using this script:
# my data:
df <- data.frame(Somatic = c("PIK3CA", "TP53", "GATA3", "KMT2C", "PTEN", "NCOR1", "ARID1A", "RUNX1", "NF1", "TBX3", "FOXA1", "ERBB2", "AKT1", "PREX2", "CBFB", "INPPL1", "NSD3", "ESR1", "AXIN2", "RAD51C"),
Freq_mut = c(32.6, 32.6, 11.9, 9.3, 5.4, 4.7, 3.8, 3.8, 3.6, 3.1, 2.9, 2.8, 2.5, 2.4, 2.3, 1, 1, 0.8, 0.6, 0.6))
library(randomcoloR)
# set of 20 random colors
colors <- distinctColorPalette(20)
ggplot(df, aes(x = "", y = Freq_mut, fill = Somatic))
geom_bar(width = 1, stat = "identity", color = "black")
coord_polar("y", start = 0)
scale_fill_manual(values = colors)
ggtitle("A")
labs(fill = "Somatic Gene")
theme(panel.background = element_blank(), plot.background = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())
geom_label_repel(data = df,
aes(y = Freq_mut, label = paste0(Somatic)),
size = 4.5, nudge_x = 1, show.legend = FALSE)
But as you can see in the image, the labels are not in the correct position and are missing the rest of the genes. How Could I fix it and remove the "x" to the left of the plot? Thanks!
CodePudding user response:
I think you may be best with direct labelling here. Remember if you have a stacked bar graph, the y values are stacked, so your text labels also need to be stacked using position_stack
library(randomcoloR)
library(geomtextpath)
# set of 20 random colors
colors <- distinctColorPalette(20)
ggplot(df, aes(x = 1, y = Freq_mut, fill = Somatic))
geom_bar(width = 1, stat = "identity", color = "black")
coord_polar("y", start = 0)
scale_fill_manual(values = colors, guide = "none")
ggtitle("A")
labs(fill = "Somatic Gene", x = NULL)
theme(panel.background = element_blank(), plot.background = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())
geom_textpath(data = df, position = position_stack(vjust = 0.5),
aes(x = 1.7, y = Freq_mut, label = paste0(Somatic)),
size = 4.5, show.legend = FALSE)
or
ggplot(df, aes(x = 1, y = Freq_mut, fill = Somatic))
geom_bar(width = 1, stat = "identity", color = "black")
coord_polar("y", start = 0)
scale_fill_manual(values = colors, guide = "none")
ggtitle("A")
labs(fill = "Somatic Gene", x = NULL)
theme(panel.background = element_blank(), plot.background = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())
geom_labelpath(data = df, position = position_stack(vjust = 0.5),
aes(x = 1.7, y = Freq_mut, label = paste0(Somatic)),
size = 4.5, show.legend = FALSE)
To add percentages, you can try:
ggplot(df, aes(x = 1, y = Freq_mut, fill = Somatic))
geom_bar(width = 1, stat = "identity", color = "black")
coord_polar("y", start = 0, clip = "off")
scale_fill_manual(values = colors, guide = "none")
xlim(0.5, 2)
ggtitle("A")
labs(fill = "Somatic Gene", x = NULL)
theme(panel.background = element_blank(), plot.background = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())
geom_textpath(data = df %>%
mutate(perc = scales::percent(Freq_mut/ sum(Freq_mut))),
position = position_stack(vjust = 0.5), hjust = 1,
aes(x = 1.55, y = Freq_mut,
label = paste0(Somatic, " (", perc, ")")),
size = 4.5, show.legend = FALSE)
CodePudding user response:
You should precalculate the positions of the labels so you can position the labels around the pie chart. Here is an option using geom_label_repel:
library(ggplot2)
library(dplyr)
library(ggrepel)
library(randomcoloR)
# set of 20 random colors
colors <- distinctColorPalette(20)
df %>%
arrange(desc(Somatic)) %>%
mutate(pos_freq_mut = cumsum(Freq_mut) - Freq_mut/2) %>%
ggplot(aes(x = "", y = Freq_mut, fill = Somatic))
geom_col(color = "black")
coord_polar("y")
geom_label_repel(aes(y = pos_freq_mut, label = paste0(Somatic)),
size = 4.5, nudge_x = 1, nudge_y = 1, show.legend = FALSE)
scale_fill_manual(values = colors)
ggtitle("A")
labs(fill = "Somatic Gene", x = "")
theme(panel.background = element_blank(), plot.background = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())