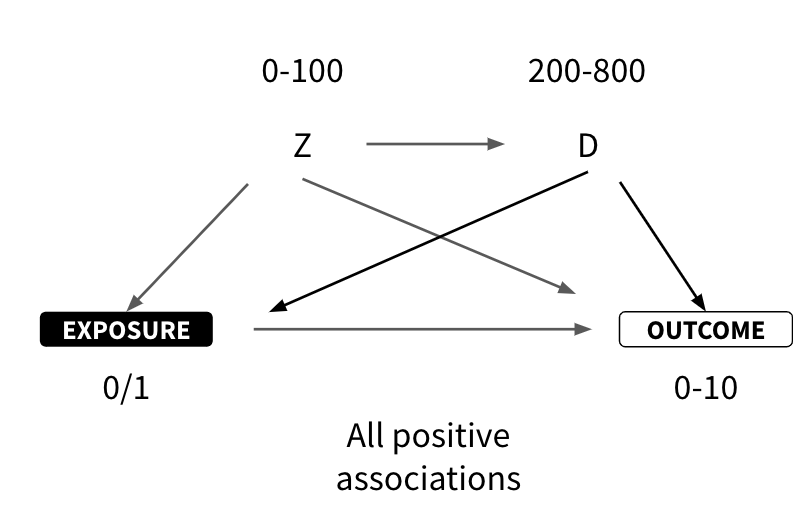
I'm looking for help simulating an observational dataset in R where the correlated variables Z and D confound the relationship between the exposure (0/1) and the outcome (0-10).
CodePudding user response:
How about something like this:
set.seed(123)
b <- replicate(2500, {
Z <- runif(250, 0, 100)
D <- scales::rescale(Z rnorm(250, 0, 20), to=c(200, 800))
q_esposure <- -5 (1/sd(Z))*Z (1/sd(D))*D
exposure <- rbinom(250, 1, plogis(q_esposure))
error <- rnorm(250, 0, 1)
outcome <- error .01*D .1*Z
outcome <- scales::rescale(outcome, to=c(0, 8.5))
outcome <- outcome 1.5*exposure
lm(outcome ~ Z D exposure)$coef
})
rowMeans(b)
#> (Intercept) Z D exposure
#> -0.722225550 0.049755855 0.004978287 1.500040770
Created on 2022-12-31 by the reprex package (v2.0.1)
CodePudding user response:
If you are looking for a single data set with the specified characteristics, you could try:
set.seed(4410)
Z <- sample(100, 1000, TRUE)
D <- Z rnorm(1000, 0, 20)
D <- round((D - min(D)) / diff(range(D)) * 600 200)
Exposure <- rbinom(1000, 1, (D Z) / 1000)
Outcome <- rbinom(1000, 10, (Exposure 2.3) / 6.6 (D Z)/2000)
df <- data.frame(Exposure, Outcome, Z, D)
Note that this gives all integers, and each of the integers is within the specified range for each variable:
head(df)
#> Exposure Outcome Z D
#> 1 0 4 8 344
#> 2 0 5 15 372
#> 3 1 7 39 549
#> 4 0 6 66 508
#> 5 1 9 78 570
#> 6 1 9 28 347
lapply(df, range)
#> $Exposure
#> [1] 0 1
#>
#> $Outcome
#> [1] 1 10
#>
#> $Z
#> [1] 1 100
#>
#> $D
#> [1] 200 800
We can confirm that Z and D are positively correlated with each other, and that both Z and D are positively correlated with Exposure:
cor(df$Z, df$D)
#> [1] 0.832381
cor(df$Z, df$Exposure)
#> [1] 0.235582
cor(df$D, df$Exposure)
#> [1] 0.2367981
Without taking the confounders into account, the effect of Exposure is about 1.8:
lm(Outcome ~ Exposure, data = df)
#>
#> Call:
#> lm(formula = Outcome ~ Exposure, data = df)
#>
#> Coefficients:
#> (Intercept) Exposure
#> 6.127 1.795
But taking the confounders into account, we can see that the coefficient for Exposure is 1.5
summary(lm(Outcome ~ Exposure Z D, data = df))
#>
#> Call:
#> lm(formula = Outcome ~ Exposure Z D, data = df)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -4.5112 -0.9401 0.0876 0.9671 4.1952
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.8034249 0.2496879 15.233 < 2e-16 ***
#> Exposure 1.5000019 0.0900666 16.654 < 2e-16 ***
#> Z 0.0035773 0.0027056 1.322 0.186
#> D 0.0045452 0.0006929 6.560 8.65e-11 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 1.364 on 996 degrees of freedom
#> Multiple R-squared: 0.378, Adjusted R-squared: 0.3761
#> F-statistic: 201.7 on 3 and 996 DF, p-value: < 2.2e-16
Created on 2022-12-31 with reprex v2.0.2