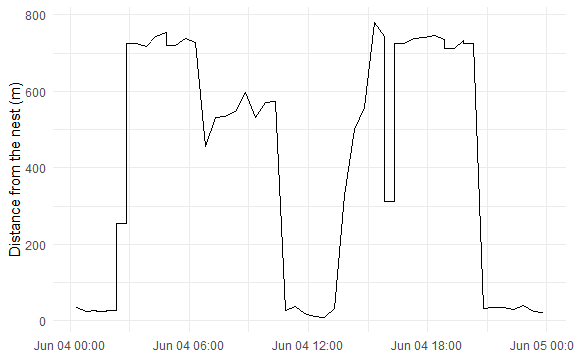
I have a large dataset, with numerous behavioral observations and I am trying to calculate the duration of each individual behaviour daily.
Here's a sample of it for 2022-06-04
library(tidyverse)
data_tibble <- tibble::tribble(
~bird_ID, ~longitude, ~latitude, ~date, ~Position_Burst_ID, ~timestamp, ~longitude_mean, ~latitude_mean, ~Dist,
"048T", 58.4836921691895, 68.8644866943359, "2022-06-04", 5121, "2022-06-04 09:49:38", 58.4966227901897, 68.8623445334237, 571.205432837044,
"048T", 58.4836730957031, 68.8645324707031, "2022-06-04", 5122, "2022-06-04 10:19:43", 58.4966227901897, 68.8623445334237, 574.044222782275,
"048T", 58.4962844848633, 68.8621215820312, "2022-06-04", 5123, "2022-06-04 10:49:36", 58.4966227901897, 68.8623445334237, 28.2914837022366,
"048T", 58.4961318969727, 68.8620452880859, "2022-06-04", 5124, "2022-06-04 11:19:44", 58.4966227901897, 68.8623445334237, 38.7040772369604,
"048T", 58.4965591430664, 68.862174987793, "2022-06-04", 5125, "2022-06-04 11:49:37", 58.4966227901897, 68.8623445334237, 19.0458862694959,
"048T", 58.4965972900391, 68.8622360229492, "2022-06-04", 5126, "2022-06-04 12:19:48", 58.4966227901897, 68.8623445334237, 12.1226274623415,
"048T", 58.4966316223145, 68.862419128418, "2022-06-04", 5127, "2022-06-04 12:49:39", 58.4966227901897, 68.8623445334237, 8.31144228341025,
"048T", 58.4971313476562, 68.8621292114258, "2022-06-04", 5128, "2022-06-04 13:19:39", 58.4966227901897, 68.8623445334237, 31.485137709381,
"048T", 58.494312286377, 68.8651657104492, "2022-06-04", 5129, "2022-06-04 13:49:41", 58.4966227901897, 68.8623445334237, 327.46020021598
)
Any idea on how to approach this?
This is the output I get for 2022-06-04:
outpout
#> # A tibble: 1 × 2
#> date duration
#> <chr> <drtn>
#> 1 2022-06-04 23.00056 hours
But I want to calculate the duration of the low points in the graph, so it should be closer to 9hrs, not 23hrs.
Edit1: I forgot to mention that I want to calculate that on a daily basis.
Edit2: I added a semi desirable output.
CodePudding user response:
Here's my approach with base function and for loop
table_recap <- data_tibble[1,]
table_recap$diff_time <- NA
tab2 <- table_recap
for (i in unique(data_tibble$bird_ID)){
subset_data_tibble <- data_tibble[data_tibble$bird_ID==i,]
subset_data_tibble <- subset_data_tibble[order(subset_data_tibble$timestamp),]
time1 <- subset_data_tibble[1,]
time2 <- subset_data_tibble[nrow(subset_data_tibble),]
table_recap$diff_time <- difftime(time2$timestamp, time1$timestamp)
tab2 <- rbind(tab2, table_recap)
}
For each "bird_ID", results will be stored in the table tab2
CodePudding user response:
Ideally, we should have sample data with multiple dates and not only for 2022-06-04
library(tidyverse)
data_tibble %>%
type_convert() %>%
group_by(date) %>%
summarise(
duration = sum(if_else(Dist <= 30, lead(timestamp) - timestamp, 0))
)
# A tibble: 1 × 2
date duration
<date> <drtn>
1 2022-06-04 120.1667 mins