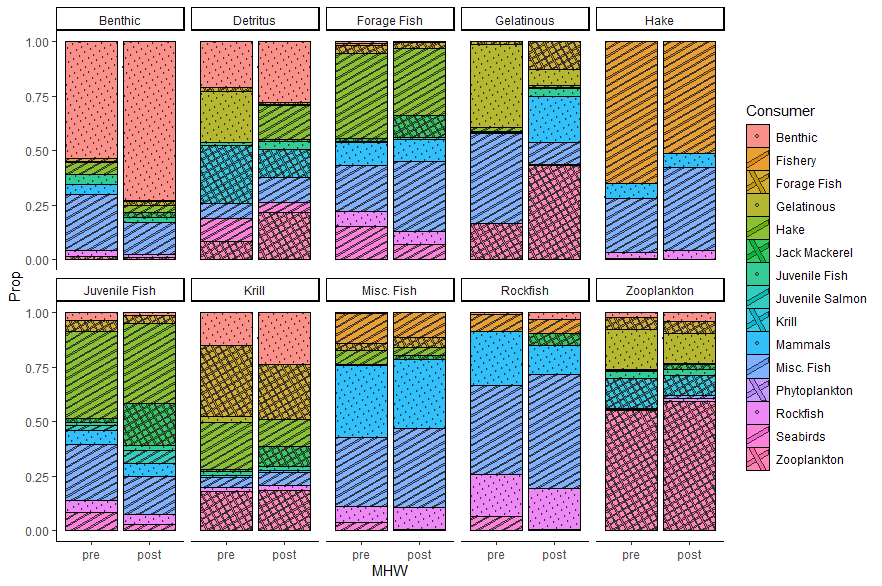
When I use the ggpattern package, it produces a great plot with a working legend in the Rstudio IDE. However, when I export to pdf or png, the legend fails to include two pattern types. Data for reproducible question at bottom of post.
# install.packages("remotes")
# remotes::install_github("coolbutuseless/ggpattern")
library(ggplot2)
library(ggpattern)
p<-ggplot(Both)
geom_col_pattern(aes(y=Prop,x=MHW,fill=Consumer,
pattern=Consumer,pattern_fill=Consumer),
color="black",alpha=0.8)
facet_wrap(~Donor,nrow=2)
theme(axis.text.x = element_text(angle=90))
theme_classic()
scale_pattern_manual(values=c(rep(c('circle','stripe', 'crosshatch'),5)))
p
Notice that all three patterns show up in the legend. Now add output to pdf or png:
pdf("Predator_Comparison/Energy_Budget_Plot.pdf",width=12,height=9)
p
dev.off()
png("Predator_Comparison/Energy_Budget_Plot.png",width=12,height=9,units = "in",res=300)
p
dev.off()
Notice that the legend now only includes the 'circle' type pattern, but the other two aren't visible. Any thoughts as to what is going on here?
There are 15 matching warnings (presumably 1 for each legend box):
Warning messages:
1: convert_polygon_sf_to_polygon_df(): Not POLYGON or MULTIPOLYGON: c("XY", "LINESTRING", "sfg")
Session Info:
> sessionInfo()
R version 4.2.2 (2022-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19044)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
attached base packages:
[1] stats graphics grDevices utils datasets methods
[7] base
other attached packages:
[1] ggpattern_1.0.0 ggplot2_3.3.6
loaded via a namespace (and not attached):
[1] Rcpp_1.0.8.3 pillar_1.7.0 compiler_4.2.2
[4] class_7.3-20 tools_4.2.2 digest_0.6.29
[7] memoise_2.0.1 lifecycle_1.0.1 tibble_3.1.7
[10] gtable_0.3.0 pkgconfig_2.0.3 rlang_1.0.6
[13] DBI_1.1.2 cli_3.3.0 rstudioapi_0.13
[16] gridpattern_1.0.1 fastmap_1.1.0 e1071_1.7-11
[19] withr_2.5.0 dplyr_1.0.9 generics_0.1.2
[22] vctrs_0.4.1 classInt_0.4-7 grid_4.2.2
[25] tidyselect_1.1.2 glue_1.6.2 sf_1.0-7
[28] R6_2.5.1 fansi_1.0.3 purrr_0.3.4
[31] farver_2.1.0 magrittr_2.0.3 units_0.8-0
[34] scales_1.2.0 ellipsis_0.3.2 assertthat_0.2.1
[37] colorspace_2.0-3 labeling_0.4.2 KernSmooth_2.23-20
[40] utf8_1.2.2 proxy_0.4-27 munsell_0.5.0
[43] cachem_1.0.6 crayon_1.5.1
Data
Both<-structure(list(Consumer = c("Phytoplankton", "Phytoplankton",
"Phytoplankton", "Phytoplankton", "Phytoplankton", "Phytoplankton",
"Phytoplankton", "Phytoplankton", "Phytoplankton", "Phytoplankton",
"Zooplankton", "Zooplankton", "Zooplankton", "Zooplankton", "Zooplankton",
"Zooplankton", "Zooplankton", "Zooplankton", "Zooplankton", "Zooplankton",
"Gelatinous", "Gelatinous", "Gelatinous", "Gelatinous", "Gelatinous",
"Gelatinous", "Gelatinous", "Gelatinous", "Gelatinous", "Gelatinous",
"Krill", "Krill", "Krill", "Krill", "Krill", "Krill", "Krill",
"Krill", "Krill", "Krill", "Forage Fish", "Forage Fish", "Forage Fish",
"Forage Fish", "Forage Fish", "Forage Fish", "Forage Fish", "Forage Fish",
"Forage Fish", "Forage Fish", "Misc. Fish", "Misc. Fish", "Misc. Fish",
"Misc. Fish", "Misc. Fish", "Misc. Fish", "Misc. Fish", "Misc. Fish",
"Misc. Fish", "Misc. Fish", "Juvenile Salmon", "Juvenile Salmon",
"Juvenile Salmon", "Juvenile Salmon", "Juvenile Salmon", "Juvenile Salmon",
"Juvenile Salmon", "Juvenile Salmon", "Juvenile Salmon", "Juvenile Salmon",
"Rockfish", "Rockfish", "Rockfish", "Rockfish", "Rockfish", "Rockfish",
"Rockfish", "Rockfish", "Rockfish", "Rockfish", "Jack Mackerel",
"Jack Mackerel", "Jack Mackerel", "Jack Mackerel", "Jack Mackerel",
"Jack Mackerel", "Jack Mackerel", "Jack Mackerel", "Jack Mackerel",
"Jack Mackerel", "Hake", "Hake", "Hake", "Hake", "Hake", "Hake",
"Hake", "Hake", "Hake", "Hake", "Juvenile Fish", "Juvenile Fish",
"Juvenile Fish", "Juvenile Fish", "Juvenile Fish", "Juvenile Fish",
"Juvenile Fish", "Juvenile Fish", "Juvenile Fish", "Juvenile Fish",
"Benthic", "Benthic", "Benthic", "Benthic", "Benthic", "Benthic",
"Benthic", "Benthic", "Benthic", "Benthic", "Seabirds", "Seabirds",
"Seabirds", "Seabirds", "Seabirds", "Seabirds", "Seabirds", "Seabirds",
"Seabirds", "Seabirds", "Mammals", "Mammals", "Mammals", "Mammals",
"Mammals", "Mammals", "Mammals", "Mammals", "Mammals", "Mammals",
"Fishery", "Fishery", "Fishery", "Fishery", "Fishery", "Fishery",
"Fishery", "Fishery", "Fishery", "Fishery", "Phytoplankton",
"Phytoplankton", "Phytoplankton", "Phytoplankton", "Phytoplankton",
"Phytoplankton", "Phytoplankton", "Phytoplankton", "Phytoplankton",
"Phytoplankton", "Zooplankton", "Zooplankton", "Zooplankton",
"Zooplankton", "Zooplankton", "Zooplankton", "Zooplankton", "Zooplankton",
"Zooplankton", "Zooplankton", "Gelatinous", "Gelatinous", "Gelatinous",
"Gelatinous", "Gelatinous", "Gelatinous", "Gelatinous", "Gelatinous",
"Gelatinous", "Gelatinous", "Krill", "Krill", "Krill", "Krill",
"Krill", "Krill", "Krill", "Krill", "Krill", "Krill", "Forage Fish",
"Forage Fish", "Forage Fish", "Forage Fish", "Forage Fish", "Forage Fish",
"Forage Fish", "Forage Fish", "Forage Fish", "Forage Fish", "Misc. Fish",
"Misc. Fish", "Misc. Fish", "Misc. Fish", "Misc. Fish", "Misc. Fish",
"Misc. Fish", "Misc. Fish", "Misc. Fish", "Misc. Fish", "Juvenile Salmon",
"Juvenile Salmon", "Juvenile Salmon", "Juvenile Salmon", "Juvenile Salmon",
"Juvenile Salmon", "Juvenile Salmon", "Juvenile Salmon", "Juvenile Salmon",
"Juvenile Salmon", "Rockfish", "Rockfish", "Rockfish", "Rockfish",
"Rockfish", "Rockfish", "Rockfish", "Rockfish", "Rockfish", "Rockfish",
"Jack Mackerel", "Jack Mackerel", "Jack Mackerel", "Jack Mackerel",
"Jack Mackerel", "Jack Mackerel", "Jack Mackerel", "Jack Mackerel",
"Jack Mackerel", "Jack Mackerel", "Hake", "Hake", "Hake", "Hake",
"Hake", "Hake", "Hake", "Hake", "Hake", "Hake", "Juvenile Fish",
"Juvenile Fish", "Juvenile Fish", "Juvenile Fish", "Juvenile Fish",
"Juvenile Fish", "Juvenile Fish", "Juvenile Fish", "Juvenile Fish",
"Juvenile Fish", "Benthic", "Benthic", "Benthic", "Benthic",
"Benthic", "Benthic", "Benthic", "Benthic", "Benthic", "Benthic",
"Seabirds", "Seabirds", "Seabirds", "Seabirds", "Seabirds", "Seabirds",
"Seabirds", "Seabirds", "Seabirds", "Seabirds", "Mammals", "Mammals",
"Mammals", "Mammals", "Mammals", "Mammals", "Mammals", "Mammals",
"Mammals", "Mammals", "Fishery", "Fishery", "Fishery", "Fishery",
"Fishery", "Fishery", "Fishery", "Fishery", "Fishery", "Fishery"
), Donor = c("Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus", "Zooplankton", "Gelatinous", "Krill", "Forage Fish",
"Misc. Fish", "Juvenile Fish", "Rockfish", "Hake", "Benthic",
"Detritus"), Prop = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0.549707229476897,
0.167643507074938, 0.177108578190667, 0, 0, 0, 0, 0, 0.0143826890253642,
0.0829032868760895, 0.187199681369435, 0.377282871954272, 0.0317762383870992,
0, 0, 0, 0, 0, 0, 0.23608971861909, 0.139938803079468, 0, 0,
0, 0, 0, 0, 0, 0, 0.26740382187952, 0.0554921943314639, 0.0141169593098825,
0.322048564282078, 0.0402201203747207, 0.0354709110190114, 0.0512084875597042,
0, 0, 0.00419945182273723, 0.0186833956330583, 0.00371942571234938,
0.413834007411829, 0.046934877854785, 0.209993132669013, 0.319007851986433,
0.258124487510548, 0.410119996920118, 0.250820097844463, 0.256537937291491,
0.0665270522130403, 0.000227877927707474, 2.19230238166815e-05,
0.000223889868244159, 0.00400793137539267, 0.000719083654571443,
0.0269857933847916, 0, 0, 0.000166958973997495, 0.000271211537513765,
0.00452310090607862, 9.0161932259385e-05, 0.0192454776488071,
0.0672607939569437, 0.0729410191750948, 0.0537692126845227, 0.191987174064799,
0.0284458999742289, 0.0285257051611284, 0, 0.00119636947274399,
4.85600349094982e-05, 0.0096441913441693, 0.0173677639651089,
0, 0.0172469482122006, 0, 0, 6.35670556389205e-05, 0.000116027210051219,
0.00429666731357137, 0.0199761039648904, 0.213659805295418, 0.387044582855991,
0.0639149633556294, 0.397293293811269, 0, 0, 0.0546098316910703,
0, 0.0316551932651675, 0.00390389142820152, 0.0172703473873986,
0, 0, 0.013396290206477, 0, 0, 0.0457699126397193, 0.0126173967159077,
0.0204905400541821, 0, 0.152368033925752, 0.00620262313406725,
0.00576668120974208, 0.0367544950174518, 0.00885310370133351,
0, 0.535495518082598, 0.207168991986137, 0.000721789625651111,
3.85712132584874e-06, 0.00072113615732755, 0.155137071890671,
0.0361919450355016, 0.0819991943267615, 0.0644369559479108, 0.00411713022515574,
2.52615601695038e-05, 0.108173638197542, 0.000831127465284076,
0.00307815674367491, 0.00899885965825438, 0.104679644151026,
0.331317128245986, 0.0632217972862738, 0.246257336982954, 0.0681378872872917,
0.0466978253528048, 4.54591320499687e-05, 0, 0, 0, 0.00808633562706585,
0.13467041631803, 0, 0.0783454323828845, 0.64847898466886, 0.0135253413432805,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0.592305587668469, 0.432622464206602,
0.185384809959524, 0, 0, 0, 0, 0, 0.0123632682828704, 0.217444293340073,
0.137061156612281, 0.0740408314288209, 0.000450958829355742,
0, 0, 0, 0, 0, 0, 0.00694691696783311, 0.0912850271659293, 0,
0, 0, 0, 0, 0, 0, 0, 0.126482511704573, 0.0572626281977687, 0.126538814376875,
0.251620851502401, 0.0293803850334056, 0.0438564810457525, 0.0398206261279957,
0, 0, 0.0193501630791624, 0.00699759838742462, 0.0133454293528453,
0.103991034069577, 0.0587719702349853, 0.323913920639485, 0.363680140068061,
0.174901149896074, 0.525917573675966, 0.380592408232399, 0.143352480934178,
0.114719759307649, 0.000716895919481306, 0.00036334504821922,
0.000827030789646068, 0.00466965533077688, 0.000145950039388543,
0.0583761853756203, 0, 0, 0.000103198641446566, 0.000667606167556876,
0.0145316852020556, 0.00277406302102769, 0.0217707477205417,
0.0567662230827084, 0.0997119449168991, 0.0474318758721208, 0.189131157773477,
0.041973522276621, 0.0134114434389902, 3.5516530117129e-05, 0.0201410109416757,
0.00341915171668221, 0.092903868588999, 0.1051958785497, 0.01838542557169,
0.194834783487864, 0.056373477474872, 0.000761098008172345, 0.0258662949582249,
0.0118196224353152, 0.00589559147923082, 0.0102581715983585,
0.1223715463388, 0.304659678104167, 0.0376880245027111, 0.365620556732433,
0, 0, 0.0304411575973729, 0.153068761838369, 0.0275540194533818,
0.0369598640935704, 0.0183898976577045, 0, 0, 0.0224593614490102,
0, 0, 0.0205323710977207, 0.0356179306572834, 0.0388436806752666,
0, 0.237769379442536, 0.000148162462610862, 0.00122497901109871,
0.0111299830615506, 0.0323568170881112, 0, 0.727171320686274,
0.27859489874831, 0.000280478691985259, 0.000223568607081643,
0.000382333975078339, 0.0711087350275364, 0.00494860881474787,
0.0272518987812531, 0.00302390077335372, 0.00169278643426349,
7.4551559576518e-06, 0.0475617036599278, 0.00077680863962975,
0.208808691833186, 0.00935660496042811, 0.102618573383442, 0.315878733012937,
0.058173579216078, 0.130457508883227, 0.0650956639065043, 0.00379652830385642,
4.28802555692741e-05, 0, 0, 0, 0.00153878838616774, 0.114479713016713,
0, 0.0627395643309926, 0.50988452114204, 0.00360431782394685,
0), MHW = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L), levels = c("pre", "post"), class = "factor")), row.names = c(NA,
-300L), class = c("tbl_df", "tbl", "data.frame"))
CodePudding user response:
Does it work if you use cairo graphics (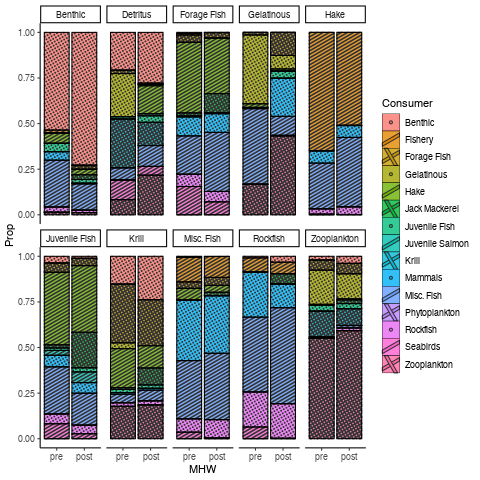
(I can't upload the pdf to SO, but the plot looks the same)
CodePudding user response: