I am working with the R programming language.
I have the following dataset:
library(dplyr)
library(purrr)
set.seed(123)
my_data1 = data.frame(var1 = rnorm(500,100,100), prop = runif(500, min=0, max=0.5))
my_data2 = data.frame(var1 = rnorm(500, 200, 50), prop = runif(500, min=0.4, max=0.7))
my_data = rbind(my_data1, my_data2)
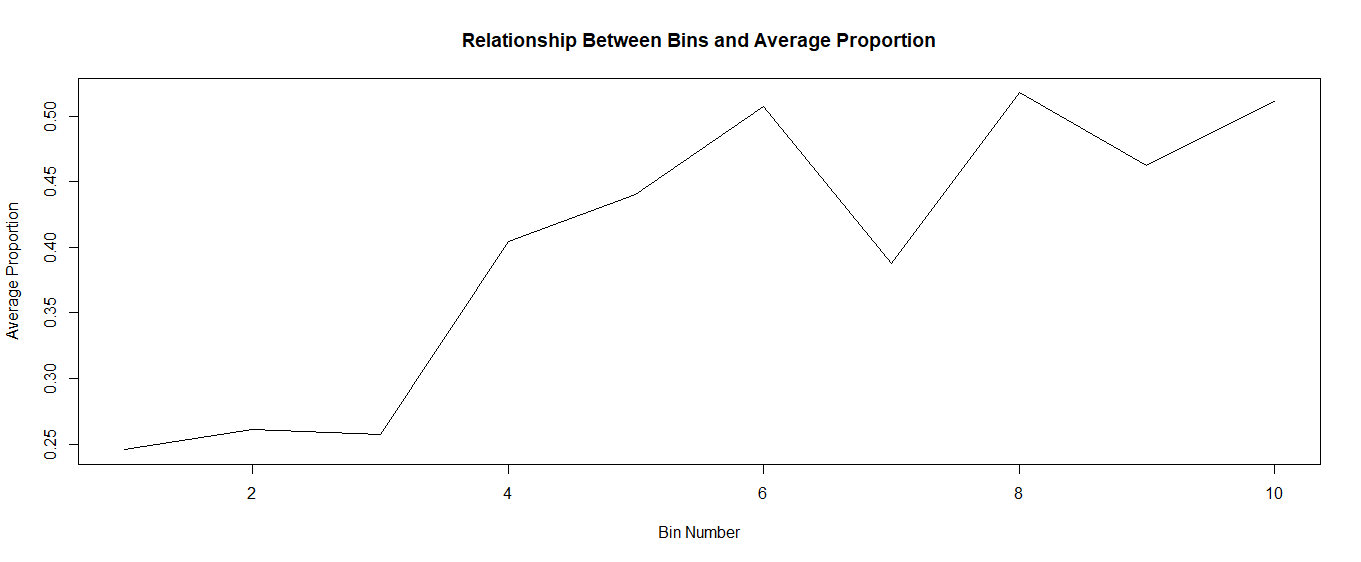
I made ten equal-sized bins, found the average proportion within each bin, then plotted the results:
final = my_data %>%
arrange(var1) %>%
mutate(ntile = ntile(var1, 10)) %>%
group_by(ntile) %>%
summarise(mean = mean(prop))
plot(final$ntile, final$mean, type = "l", xlab = "Bin Number", ylab = "Average Proportion", main = "Relationship Between Bins and Average Proportion")
Now, suppose I:
- Take a random 70% sample, create 10 bins and calculate the mean proportion within each of these bins
- Then, from the remaining 30% sample - I again take a random sample and create 10 bins and calculate the mean proportion within each of the bins
- Next, I take the squared sum of the difference between these two steps for each bin
- Finally, repeat this process many times (while tracking the results)
Below, I wrote some R code for this procedure:
base = sample_n(my_data, 700)
base_comp = base %>%
arrange(var1) %>%
mutate(ntile = ntile(var1, 10)) %>%
group_by(ntile) %>%
summarise(mean = mean(prop))
sampling_frame = my_data %>% anti_join(base)
my_list = list()
for (i in 1:1000)
{
a_i = sample_n(sampling_frame, 100)
base_a_i = a_i %>%
arrange(var1) %>%
mutate(ntile = ntile(var1, 10)) %>%
group_by(ntile) %>%
summarise(mean = mean(prop))
sum_i <- sum(map2_dbl(base_a_i$mean, base_comp$mean, function(x, y) (x - y)^2))
my_list[[i]] <- data.frame(id = i, sum_i)
print(data.frame(id = i, sum_i))
}
results = do.call(rbind.data.frame, my_list)
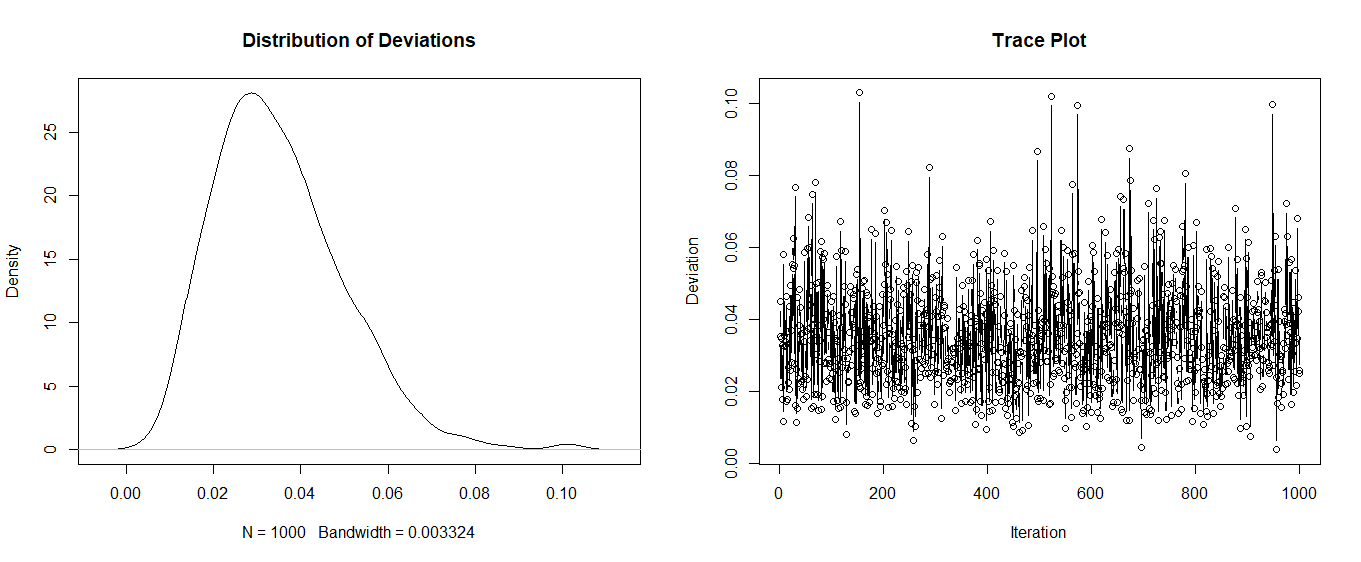
plot(density(results$sum_i), main = "Distribution of Deviations")
plot(results$id, results$sum_i, xlab = "Iteration", ylab = "Deviation", main = "Trace Plot", type = "b")
My Question: Now, I am trying to create a "double loop" - that is, I would to:
- For j = 1, randomly create "base_j" , "base_comp_j" and "sampling_frame_j"
- Repeat the "i" loop 100 times
- Take the average of all "sum_i" for j=1 (i.e. average_sum_i_j)
- Now, repeat this for j = 2
- Repeat until j = 100
- Each point on these two graphs will be "average_sum_i_j"
Here is my attempt to do this:
# change i and j index to 10 for brevity
my_list = list()
for (j in 1:10) {
base_j = sample_n(my_data, 700)
base_comp_j = base_j %>%
arrange(var1) %>%
mutate(ntile = ntile(var1, 10)) %>%
group_by(ntile) %>%
summarise(mean = mean(prop))
sampling_frame_j = my_data %>% anti_join(base_j)
sum_i_list = list()
for (i in 1:10) {
a_i = sample_n(sampling_frame_j, 100)
base_a_i = a_i %>%
arrange(var1) %>%
mutate(ntile = ntile(var1, 10)) %>%
group_by(ntile) %>%
summarise(mean = mean(prop))
sum_i <- sum(map2_dbl(base_a_i$mean, base_comp_j$mean, function(x, y) (x - y)^2))
sum_i_list[[i]] <- sum_i
}
average_sum_i_j = mean(unlist(sum_i_list))
my_list[[j]] <- data.frame(id_j = j, id_i = i, average_sum_i_j)
print(data.frame(id_j = j, id_i = i, average_sum_i_j))
}
results = do.call(rbind.data.frame, my_list)
plot(density(results$average_sum_i_j), main = "Distribution of Deviations")
plot(results$id, results$average_sum_i_j, xlab = "Iteration", ylab = "Deviation", main = "Trace Plot", type = "b")
However, I don't think I am doing this right since the loop does not appear to be cycling through both indices.
Can someone show me how to correct this?
Thanks!
CodePudding user response:
Here is the question's code revised to correct some, not many, problems it has.
I have commented the changes in the code, the rest should be self-explanatory.
I have also commented out the data.frame printing.
suppressPackageStartupMessages({
library(dplyr)
library(purrr)
})
set.seed(123)
my_data1 = data.frame(var1 = rnorm(500,100,100), prop = runif(500, min=0, max=0.5))
my_data2 = data.frame(var1 = rnorm(500, 200, 50), prop = runif(500, min=0.4, max=0.7))
my_data = rbind(my_data1, my_data2)
# define these constants, it will be easier to change
# them here than in the loops
nbins <- 10L
n_loop_i <- 10L
n_loop_j <- 100L
n_my_data <- nrow(my_data)
sampling_size_j <- 700L
sampling_size_i <- 100L
# create the results list beforehand, much more memory
# and speed efficient
my_list <- vector("list", length = n_loop_j)
for (j in seq.int(n_loop_j)) {
#
# avoids an expensive call to anti_join
# all that is needed is, after the pipe,
# to negate the index jnx
jnx <- sample(n_my_data, sampling_size_j)
base_j <- my_data[jnx, ]
base_comp_j <- base_j %>%
arrange(var1) %>%
mutate(ntile = ntile(var1, nbins)) %>%
group_by(ntile) %>%
summarise(mean = mean(prop))
sampling_frame_j <- my_data[-jnx, ]
#
# it's simpler to create and populate a vector
# than a list and after the i loop it will
# avoid a function call (unlist)
sum_i_vec <- numeric(n_loop_i)
for (i in seq.int(n_loop_i)) {
a_i <- sample_n(sampling_frame_j, sampling_size_i)
base_a_i <- a_i %>%
arrange(var1) %>%
mutate(ntile = ntile(var1, 10)) %>%
group_by(ntile) %>%
summarise(mean = mean(prop))
sum_i <- sum(map2_dbl(base_a_i$mean, base_comp_j$mean, function(x, y) (x - y)^2))
sum_i_vec[i] <- sum_i
}
average_sum_i_j <- mean(sum_i_vec)
#
# I would get rid of id_i = i, it is always the
# last value of seq.int(n_loop_i)
my_list[[j]] <- data.frame(id_j = j, id_i = i, average_sum_i_j)
#print(data.frame(id_j = j, id_i = i, average_sum_i_j))
}
results <- do.call(rbind.data.frame, my_list)
str(results)
#> 'data.frame': 100 obs. of 3 variables:
#> $ id_j : int 1 2 3 4 5 6 7 8 9 10 ...
#> $ id_i : int 10 10 10 10 10 10 10 10 10 10 ...
#> $ average_sum_i_j: num 0.0522 0.0266 0.0494 0.0278 0.0244 ...
# save current graphic settings
old_par <- par(mfrow = c(1, 2))
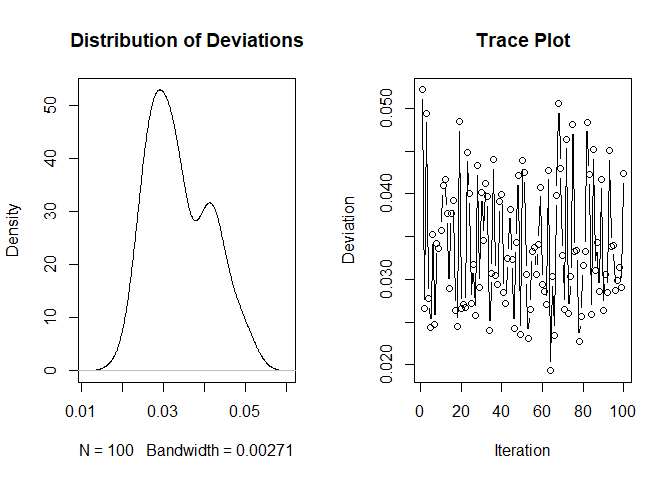
plot(density(results$average_sum_i_j), main = "Distribution of Deviations")
plot(results$id_j, results$average_sum_i_j,
xlab = "Iteration", ylab = "Deviation",
main = "Trace Plot", type = "b")
# restore graphic settings
par(old_par)
Created on 2023-01-20 with reprex v2.0.2