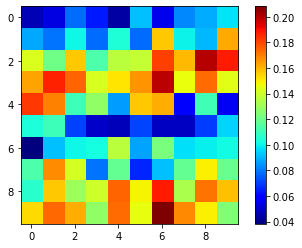
I have a 10 by 10 numpy array. For every cell in the array (as depicted in the snapshot below), I wanted to make calculations based on left, right, top, and bottom neighboring cells, as well as a weighted moving average window based on the surrounding grid cells around each cell in the numpy array (based on its i and j index). I was not sure how to account for grid cells that are located at the edges (boundaries and corners, as they only have a partial of the window kernel around them). Is there any pythonic way to perform these types of tasks? For the weighted_moving_average, the 8 immediate surrounding cells should have a 70% (stronger) whereas the not_immediate sorounding cells (12) should have 30% weights (weaker).
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
noisy_data = [[0.0467251, 0.0529133, 0.0775945, 0.0640082, 0.0439774, 0.0915829,
0.0547699, 0.0826791, 0.0883616, 0.0977739],
[0.0875183, 0.0790368, 0.101146 , 0.0777582, 0.104289 , 0.0775479,
0.156569 , 0.0999909, 0.0905242, 0.161871 ],
[0.1427 , 0.1211 , 0.15661 , 0.113337 , 0.135986 , 0.1384 ,
0.181515 , 0.159102 , 0.200004 , 0.187535 ],
[0.163381 , 0.186425 , 0.174798 , 0.14191 , 0.1514 , 0.166044 ,
0.199063 , 0.146319 , 0.173303 , 0.144067 ],
[0.182485 , 0.16901 , 0.11 , 0.127162 , 0.0858656, 0.1564 ,
0.161187 , 0.0588985, 0.110706 , 0.0560944],
[0.103804 , 0.109948 , 0.0707144, 0.0493536, 0.0471329, 0.0708205,
0.048201 , 0.0487304, 0.069967 , 0.0947939],
[0.0382397, 0.0918 , 0.101197 , 0.102453 , 0.135943 , 0.0867575,
0.123071 , 0.0970329, 0.099906 , 0.101536 ],
[0.113361 , 0.167597 , 0.141526 , 0.0792396, 0.118 , 0.065764 ,
0.0915817, 0.1183 , 0.149376 , 0.118608 ],
[0.10647 , 0.1566 , 0.1303 , 0.139754 , 0.174465 , 0.148458 ,
0.187625 , 0.132805 , 0.171687 , 0.158161 ],
[0.15319 , 0.174204 , 0.161628 , 0.127377 , 0.173368 , 0.145292 ,
0.208068 , 0.168076 , 0.14961 , 0.124334 ]]
noisy_data = np.array(noisy_data).reshape(10, 10)
plt.imshow(noisy_data, cmap ="jet")
plt.colorbar()
new_df = pd.DataFrame()
new_df['left_neighboring_index') = [(np.abs(noisy_data[i-1] -noisy_data[i]))/(noisy_data[i-1) noisy_data[i]) for i in noisy_data]
new_df['right_neighboring_index') = [(np.abs(noisy_data[i 1] -noisy_data[i]))/(noisy_data[i 1) noisy_data[i]) for i in noisy_data]
new_df['bottom_neighboring_index') = [(np.abs(noisy_data[j-1] -noisy_data[j]))/(noisy_data[j-1) noisy_data[j]) for i in noisy_data]
new_df['top_neighboring_index') = [(np.abs(noisy_data[j 1] -noisy_data[j]))/(noisy_data[j 1) noisy_data[j]) for i in noisy_data]
new_df['weighted_moving_average_size_20_(8 12)') = ...?
CodePudding user response:
I think what you're looking for is what is known as a convolution. In a convolution on a matrix, a kernel (smaller matrix) is swept across the matrix.
I can try to explain this but I rather suggest you watch this 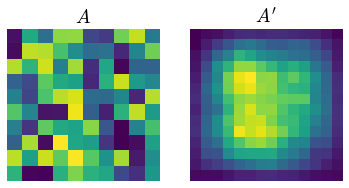
There are several ways to deal with boundary cells (aslo explained in the video I think). For this you can use the boundary argument in the convolve2D function (see documentation).
CodePudding user response:
You are trying to do operation similar to 2D convolution but not exactly convolution. In convolution, we do multiply with a kernel and then sum the resulting value and place it in each cell. You want the average of the product to be placed in the cell instead.
def surr_avg(img, kernel):
kernel_shape = kernel.shape
# get the sliding window view of the image.
sub_matrices = np.lib.stride_tricks.sliding_window_view(img, kernel_shape)
# Multiply kernel with each window of img using einsum.
conv = np.einsum('kl,ijkl->ijkl',kernel,sub_matrices)
# find the reduced mean along the last two axis.
return conv.mean(axis=(-1,-2))
For simple average:
k = np.array([[1,1,1],
[1,0,1],
[1,1,1]])
avg = surr_avg(a, k)
For weighted average:
wk = np.array([
[.3, .3, .3, .3, .3],
[.3, .7, .7, .7, .3],
[.3, .7, .0, .7, .3],
[.3, .7, .7, .7, .3],
[.3, .3, .3, .3, .3]
])
wk /= wk.sum()
wght_avg = surr_avg(a, wk)