I have a dataset consist of 4 variables: CR, EN, LC and VU:
View first few values of my dateset
CR = c(2, 9, 10, 14, 24, 27, 29, 30, 34, 43, 50, 74, 86, 105, 140, 155, 200, …)
EN = c(24, 52, 86, 110, 144, 154, 206, 242, 300, 302, 366, 403, 422, 427, 427, 434, 448, …)
LC = c(447, 476, 543, 580, 647, 685, 745, 763, 819, 821, 863, 904, 908, 926, 934, 951, 968, …)
VU = c(75, 96, 97, 217, 297, 498, 511, 551, 560, 564, 570, 575, 609, 673, 681, 700, 755,...)
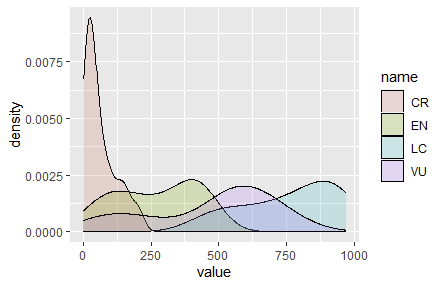
I want to create a histogram of a group of these variables in a plot by R that shows the normal distribution and density, a plot similar to the one below...
CodePudding user response:
Here are the distributions, a clear-cut use of geom_density.
But first, to address "grouping", we need to pivot/reshape the data so that ggplot2 can automatically handle grouping. This will result in a column with a character (or factor) for each of the "CR", "EN", "LC", or "VU", and another column with the particular value. When pivoting, there is typically one or more columns that are preserved (an id, an x-value, a time/date, or something similar), but we don't have any data that would suggest something to preserve.
longdat <- tidyr::pivot_longer(dat, everything())
longdat
# # A tibble: 68 × 2
# name value
# <chr> <dbl>
# 1 CR 2
# 2 EN 24
# 3 LC 447
# 4 VU 75
# 5 CR 9
# 6 EN 52
# 7 LC 476
# 8 VU 96
# 9 CR 10
# 10 EN 86
# # … with 58 more rows
# # ℹ Use `print(n = ...)` to see more rows
ggplot(longdat, aes(x = value, group = name, fill = name))
geom_density(alpha = 0.2)
tidyr::pivot_longer works, one can also use melt from either reshape2 or data.table:
longdat <- reshape2::melt(dat, c())
## names are 'variable' and 'value' instead of 'name' and 'value'
Data
dat <- structure(list(CR = c(2, 9, 10, 14, 24, 27, 29, 30, 34, 43, 50, 74, 86, 105, 140, 155, 200), EN = c(24, 52, 86, 110, 144, 154, 206, 242, 300, 302, 366, 403, 422, 427, 427, 434, 448), LC = c(447, 476, 543, 580, 647, 685, 745, 763, 819, 821, 863, 904, 908, 926, 934, 951, 968), VU = c(75, 96, 97, 217, 297, 498, 511, 551, 560, 564, 570, 575, 609, 673, 681, 700, 755)), class = "data.frame", row.names = c(NA, -17L))