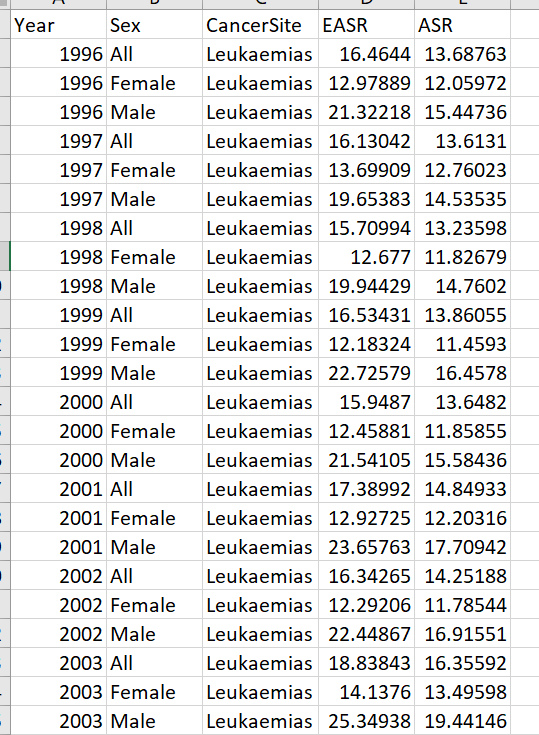
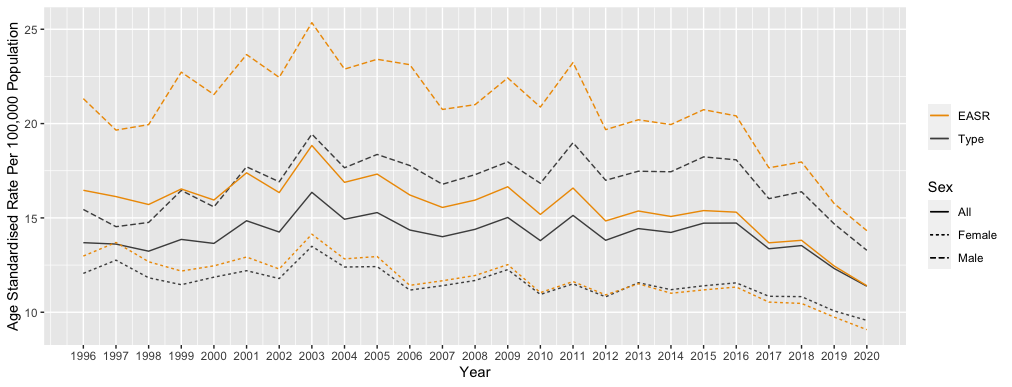
I'm trying to produce a line plot with 2 y axes. I would like to include groups for each Y axis (i.e., Male, Female, All). There should be 6 lines on the plot in total. EASR on AXis 1 and ASR on Axis 2 with Year as the x axis.
plt_EASR_ASR = ggplot(EASR_ASR, aes(x = Year, y = c_rate_AllAges))
geom_line(aes(color = "Type"))
geom_line(aes(y = EASR, color = "EASR"))
scale_x_continuous(breaks = seq(1996,2020,by=1))
labs(x = "Year", y = "Age Standardised Rate Per 100,000 Population", color = "")
scale_color_manual(values = c("orange2", "gray30"))
This is the code I've tried but I can't figure out how to get groups within each Y axis
Any help would be greatly appreciated.
"Year","Sex","CancerSite","EASR","c_rate_AllAges" 1996,"All","Leukaemias",16.4644014652654,13.6876275237177 1996,"Female","Leukaemias",12.9788937078698,12.0597163887387 1996,"Male","Leukaemias",21.3221816440295,15.4473604629304 1997,"All","Leukaemias",16.1304181900654,13.6130969008565 1997,"Female","Leukaemias",13.6990871096256,12.7602310927074 1997,"Male","Leukaemias",19.6538259658305,14.5353479189067 1998,"All","Leukaemias",15.7099429183033,13.235980595107 1998,"Female","Leukaemias",12.6770034224731,11.8267875980969 1998,"Male","Leukaemias",19.944293692904,14.7602020671663 1999,"All","Leukaemias",16.5343082389775,13.8605467325191 1999,"Female","Leukaemias",12.1832360670675,11.4592957845349 1999,"Male","Leukaemias",22.725789023191,16.4577976511727 2000,"All","Leukaemias",15.9486981922539,13.6481965024274 2000,"Female","Leukaemias",12.4588080610428,11.8585457926108 2000,"Male","Leukaemias",21.5410513177349,15.5843557739833 2001,"All","Leukaemias",17.3899183283119,14.8493345444493 2001,"Female","Leukaemias",12.9272492377534,12.2031563216722 2001,"Male","Leukaemias",23.6576342297075,17.7094200555279 2002,"All","Leukaemias",16.342652248546,14.251875246743 2002,"Female","Leukaemias",12.2920630166641,11.7854366219137 2002,"Male","Leukaemias",22.4486690429546,16.9155066337937 2003,"All","Leukaemias",18.8384318858525,16.3559238433462 2003,"Female","Leukaemias",14.1376004765121,13.4959846594374 2003,"Male","Leukaemias",25.3493810717121,19.4414639668462 2004,"All","Leukaemias",16.8847449806357,14.928308715064 2004,"Female","Leukaemias",12.8283318157582,12.3971782960666 2004,"Male","Leukaemias",22.887108143603,17.6571352197312 2005,"All","Leukaemias",17.3223262286823,15.2831591718524 2005,"Female","Leukaemias",12.953461245947,12.4201280065959 2005,"Male","Leukaemias",23.4074022185601,18.3644730330715 2006,"All","Leukaemias",16.2159145501269,14.3580751996883 2006,"Female","Leukaemias",11.4258263330529,11.1740839508551 2006,"Male","Leukaemias",23.1230246136623,17.7773180814718 2007,"All","Leukaemias",15.5548363062569,14.0038684719536 2007,"Female","Leukaemias",11.6685220505893,11.4085180858676 2007,"Male","Leukaemias",20.7487826782262,16.7831002591968 2008,"All","Leukaemias",15.9445080762995,14.3958177170424 2008,"Female","Leukaemias",11.9466644838843,11.6832775090955 2008,"Male","Leukaemias",21.0028913088726,17.2941734935582 2009,"All","Leukaemias",16.6510295938606,15.0232229209274 2009,"Female","Leukaemias",12.5247418708032,12.2595725594469 2009,"Male","Leukaemias",22.4208474149683,17.9702042165603 2010,"All","Leukaemias",15.1844362788918,13.7965109649956 2010,"Female","Leukaemias",11.0402986296808,10.943329764162 2010,"Male","Leukaemias",20.8759969211269,16.8352943115542 2011,"All","Leukaemias",16.5825212501415,15.1323609879432 2011,"Female","Leukaemias",11.6265977143274,11.5035169988277 2011,"Male","Leukaemias",23.2356253141588,18.9861105707505 2012,"All","Leukaemias",14.8398166311995,13.8136103583258 2012,"Female","Leukaemias",10.917884698992,10.8174877846443 2012,"Male","Leukaemias",19.678158008849,16.9945950979517 2013,"All","Leukaemias",15.3664342290484,14.433995908178 2013,"Female","Leukaemias",11.5092416333904,11.5650378326316 2013,"Male","Leukaemias",20.2009806078599,17.474136731254 2014,"All","Leukaemias",15.0768981887139,14.2306829231805 2014,"Female","Leukaemias",11.0073849526538,11.1956438767461 2014,"Male","Leukaemias",19.9480316140044,17.4463611050132 2015,"All","Leukaemias",15.3885098810921,14.7217569328122 2015,"Female","Leukaemias",11.1836648830556,11.4025869754946 2015,"Male","Leukaemias",20.7368901755758,18.2342713129327 2016,"All","Leukaemias",15.304270816888,14.7279219938202 2016,"Female","Leukaemias",11.3364493262975,11.5584166337498 2016,"Male","Leukaemias",20.4067238009368,18.0780002915315 2017,"All","Leukaemias",13.6796869098636,13.3645480017697 2017,"Female","Leukaemias",10.5377741734033,10.8457532770695 2017,"Male","Leukaemias",17.6552137663892,16.020906715146 2018,"All","Leukaemias",13.8134223485237,13.5341387616999 2018,"Female","Leukaemias",10.4671825211221,10.8268990362984 2018,"Male","Leukaemias",17.9647211234894,16.3850811193653 2019,"All","Leukaemias",12.4496132202679,12.3185620412571 2019,"Female","Leukaemias",9.74267026583603,10.0703603939154 2019,"Male","Leukaemias",15.7610080489667,14.6826721562086 2020,"All","Leukaemias",11.4067274612956,11.3794365166484 2020,"Female","Leukaemias",9.07667268404083,9.56873565582258 2020,"Male","Leukaemias",14.3256054709995,13.2822454649011
CodePudding user response:
Perhaps you want to map Sex to an aesthetic to separate the series?
ggplot(EASR_ASR, aes(x = Year, y = c_rate_AllAges))
geom_line(aes(color = "Type", linetype = Sex))
geom_line(aes(y = EASR, color = "EASR", linetype = Sex))
scale_x_continuous(breaks = seq(1996,2020,by=1))
labs(x = "Year", y = "Age Standardised Rate Per 100,000 Population", color = "")
scale_color_manual(values = c("orange2", "gray30"))
Example data
EASR_ASR <- data.frame(
stringsAsFactors = FALSE,
Year = c(1996L,
1996L,1996L,1997L,1997L,1997L,1998L,
1998L,1998L,1999L,1999L,1999L,2000L,2000L,
2000L,2001L,2001L,2001L,2002L,2002L,
2002L,2003L,2003L,2003L,2004L,2004L,2004L,
2005L,2005L,2005L,2006L,2006L,2006L,
2007L,2007L,2007L,2008L,2008L,2008L,
2009L,2009L,2009L,2010L,2010L,2010L,2011L,
2011L,2011L,2012L,2012L,2012L,2013L,
2013L,2013L,2014L,2014L,2014L,2015L,2015L,
2015L,2016L,2016L,2016L,2017L,2017L,
2017L,2018L,2018L,2018L,2019L,2019L,2019L,
2020L,2020L,2020L),
Sex = c("All",
"Female","Male","All","Female","Male",
"All","Female","Male","All","Female",
"Male","All","Female","Male","All","Female",
"Male","All","Female","Male","All",
"Female","Male","All","Female","Male","All",
"Female","Male","All","Female","Male",
"All","Female","Male","All","Female",
"Male","All","Female","Male","All","Female",
"Male","All","Female","Male","All",
"Female","Male","All","Female","Male","All",
"Female","Male","All","Female","Male",
"All","Female","Male","All","Female",
"Male","All","Female","Male","All","Female",
"Male","All","Female","Male"),
CancerSite = c("Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias","Leukaemias","Leukaemias",
"Leukaemias"),
EASR = c(16.46440147,12.97889371,21.32218164,16.13041819,
13.69908711,19.65382597,15.70994292,
12.67700342,19.94429369,16.53430824,12.18323607,
22.72578902,15.94869819,12.45880806,
21.54105132,17.38991833,12.92724924,23.65763423,
16.34265225,12.29206302,22.44866904,
18.83843189,14.13760048,25.34938107,
16.88474498,12.82833182,22.88710814,17.32232623,
12.95346125,23.40740222,16.21591455,
11.42582633,23.12302461,15.55483631,11.66852205,
20.74878268,15.94450808,11.94666448,
21.00289131,16.65102959,12.52474187,22.42084741,
15.18443628,11.04029863,20.87599692,
16.58252125,11.62659771,23.23562531,14.83981663,
10.9178847,19.67815801,15.36643423,
11.50924163,20.20098061,15.07689819,11.00738495,
19.94803161,15.38850988,11.18366488,
20.73689018,15.30427082,11.33644933,20.4067238,
13.67968691,10.53777417,17.65521377,
13.81342235,10.46718252,17.96472112,12.44961322,
9.742670266,15.76100805,11.40672746,
9.076672684,14.32560547),
c_rate_AllAges = c(13.68762752,12.05971639,15.44736046,13.6130969,
12.76023109,14.53534792,13.2359806,
11.8267876,14.76020207,13.86054673,11.45929578,
16.45779765,13.6481965,11.85854579,
15.58435577,14.84933454,12.20315632,17.70942006,
14.25187525,11.78543662,16.91550663,
16.35592384,13.49598466,19.44146397,14.92830872,
12.3971783,17.65713522,15.28315917,
12.42012801,18.36447303,14.3580752,11.17408395,
17.77731808,14.00386847,11.40851809,
16.78310026,14.39581772,11.68327751,17.29417349,
15.02322292,12.25957256,17.97020422,
13.79651096,10.94332976,16.83529431,15.13236099,
11.503517,18.98611057,13.81361036,
10.81748778,16.9945951,14.43399591,11.56503783,
17.47413673,14.23068292,11.19564388,
17.44636111,14.72175693,11.40258698,18.23427131,
14.72792199,11.55841663,18.07800029,
13.364548,10.84575328,16.02090672,13.53413876,
10.82689904,16.38508112,12.31856204,
10.07036039,14.68267216,11.37943652,9.568735656,
13.28224546)
)
CodePudding user response:
Mapping Sex to an aesthetic is probably what you want, but as an exercise, here's how to get closer to recreating the plot above.
library(dplyr)
library(tidyr)
library(ggplot2)
EASR_ASR <- tibble::tribble(
~"Year",~"Sex",~"CancerSite",~"EASR",~"c_rate_AllAges",
1996,"All","Leukaemias",16.4644014652654,13.6876275237177,
1996,"Female","Leukaemias",12.9788937078698,12.0597163887387,
1996,"Male","Leukaemias",21.3221816440295,15.4473604629304,
1997,"All","Leukaemias",16.1304181900654,13.6130969008565,
1997,"Female","Leukaemias",13.6990871096256,12.7602310927074,
1997,"Male","Leukaemias",19.6538259658305,14.5353479189067,
1998,"All","Leukaemias",15.7099429183033,13.235980595107,
1998,"Female","Leukaemias",12.6770034224731,11.8267875980969,
1998,"Male","Leukaemias",19.944293692904,14.7602020671663,
1999,"All","Leukaemias",16.5343082389775,13.8605467325191,
1999,"Female","Leukaemias",12.1832360670675,11.4592957845349,
1999,"Male","Leukaemias",22.725789023191,16.4577976511727,
2000,"All","Leukaemias",15.9486981922539,13.6481965024274,
2000,"Female","Leukaemias",12.4588080610428,11.8585457926108,
2000,"Male","Leukaemias",21.5410513177349,15.5843557739833,
2001,"All","Leukaemias",17.3899183283119,14.8493345444493,
2001,"Female","Leukaemias",12.9272492377534,12.2031563216722,
2001,"Male","Leukaemias",23.6576342297075,17.7094200555279,
2002,"All","Leukaemias",16.342652248546,14.251875246743,
2002,"Female","Leukaemias",12.2920630166641,11.7854366219137,
2002,"Male","Leukaemias",22.4486690429546,16.9155066337937,
2003,"All","Leukaemias",18.8384318858525,16.3559238433462,
2003,"Female","Leukaemias",14.1376004765121,13.4959846594374,
2003,"Male","Leukaemias",25.3493810717121,19.4414639668462,
2004,"All","Leukaemias",16.8847449806357,14.928308715064,
2004,"Female","Leukaemias",12.8283318157582,12.3971782960666,
2004,"Male","Leukaemias",22.887108143603,17.6571352197312,
2005,"All","Leukaemias",17.3223262286823,15.2831591718524,
2005,"Female","Leukaemias",12.953461245947,12.4201280065959,
2005,"Male","Leukaemias",23.4074022185601,18.3644730330715,
2006,"All","Leukaemias",16.2159145501269,14.3580751996883,
2006,"Female","Leukaemias",11.4258263330529,11.1740839508551,
2006,"Male","Leukaemias",23.1230246136623,17.7773180814718,
2007,"All","Leukaemias",15.5548363062569,14.0038684719536,
2007,"Female","Leukaemias",11.6685220505893,11.4085180858676,
2007,"Male","Leukaemias",20.7487826782262,16.7831002591968,
2008,"All","Leukaemias",15.9445080762995,14.3958177170424,
2008,"Female","Leukaemias",11.9466644838843,11.6832775090955,
2008,"Male","Leukaemias",21.0028913088726,17.2941734935582,
2009,"All","Leukaemias",16.6510295938606,15.0232229209274,
2009,"Female","Leukaemias",12.5247418708032,12.2595725594469,
2009,"Male","Leukaemias",22.4208474149683,17.9702042165603,
2010,"All","Leukaemias",15.1844362788918,13.7965109649956,
2010,"Female","Leukaemias",11.0402986296808,10.943329764162,
2010,"Male","Leukaemias",20.8759969211269,16.8352943115542,
2011,"All","Leukaemias",16.5825212501415,15.1323609879432,
2011,"Female","Leukaemias",11.6265977143274,11.5035169988277,
2011,"Male","Leukaemias",23.2356253141588,18.9861105707505,
2012,"All","Leukaemias",14.8398166311995,13.8136103583258,
2012,"Female","Leukaemias",10.917884698992,10.8174877846443,
2012,"Male","Leukaemias",19.678158008849,16.9945950979517,
2013,"All","Leukaemias",15.3664342290484,14.433995908178,
2013,"Female","Leukaemias",11.5092416333904,11.5650378326316,
2013,"Male","Leukaemias",20.2009806078599,17.474136731254,
2014,"All","Leukaemias",15.0768981887139,14.2306829231805,
2014,"Female","Leukaemias",11.0073849526538,11.1956438767461,
2014,"Male","Leukaemias",19.9480316140044,17.4463611050132,
2015,"All","Leukaemias",15.3885098810921,14.7217569328122,
2015,"Female","Leukaemias",11.1836648830556,11.4025869754946,
2015,"Male","Leukaemias",20.7368901755758,18.2342713129327,
2016,"All","Leukaemias",15.304270816888,14.7279219938202,
2016,"Female","Leukaemias",11.3364493262975,11.5584166337498,
2016,"Male","Leukaemias",20.4067238009368,18.0780002915315,
2017,"All","Leukaemias",13.6796869098636,13.3645480017697,
2017,"Female","Leukaemias",10.5377741734033,10.8457532770695,
2017,"Male","Leukaemias",17.6552137663892,16.020906715146,
2018,"All","Leukaemias",13.8134223485237,13.5341387616999,
2018,"Female","Leukaemias",10.4671825211221,10.8268990362984,
2018,"Male","Leukaemias",17.9647211234894,16.3850811193653,
2019,"All","Leukaemias",12.4496132202679,12.3185620412571,
2019,"Female","Leukaemias",9.74267026583603,10.0703603939154,
2019,"Male","Leukaemias",15.7610080489667,14.6826721562086,
2020,"All","Leukaemias",11.4067274612956,11.3794365166484,
2020,"Female","Leukaemias",9.07667268404083,9.56873565582258,
2020,"Male","Leukaemias",14.3256054709995,13.2822454649011)
mxdat <- EASR_ASR %>% filter(Sex!= "All") %>% select(4:5) %>% summarise(across(everything(), max))
rs_fac <- mxdat$c_rate_AllAges/mxdat$EASR
inv_rs_fac <- mxdat$EASR/mxdat$c_rate_AllAges
tmp <- EASR_ASR %>%
filter(Sex %in% c("Male", "Female")) %>%
mutate(EASR = EASR*rs_fac) %>%
pivot_longer(4:5, names_to="vbl", values_to="vals") %>%
rowwise() %>%
mutate(group = paste(Sex, vbl, sep=": ")) %>%
ungroup()
ggplot(tmp, aes(x = Year, y = vals, colour=group, linetype=group))
geom_line()
scale_colour_manual(values=c("orange2", "orange2", "gray30", "gray30"))
scale_linetype_manual(values=c(1,3,1,3))
scale_x_continuous(breaks = seq(1996,2020,by=1))
scale_y_continuous(limits = c(0,20),
sec.axis = sec_axis(trans=~.x*inv_rs_fac,
breaks = round(c(0,5,10,15,20)*inv_rs_fac, 1), name = "EASR"))
labs(x = "Year", y = "Age Standardised Rate Per 100,000 Population",
color = "", linetype="")
theme_bw()
theme(axis.text.x=element_text(angle=45, hjust=1),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank(),
panel.grid.minor.y = element_blank(),
legend.position = c(.5, .15),
legend.justification = "center",
legend.background = element_blank())
guides(col=guide_legend(ncol=2))
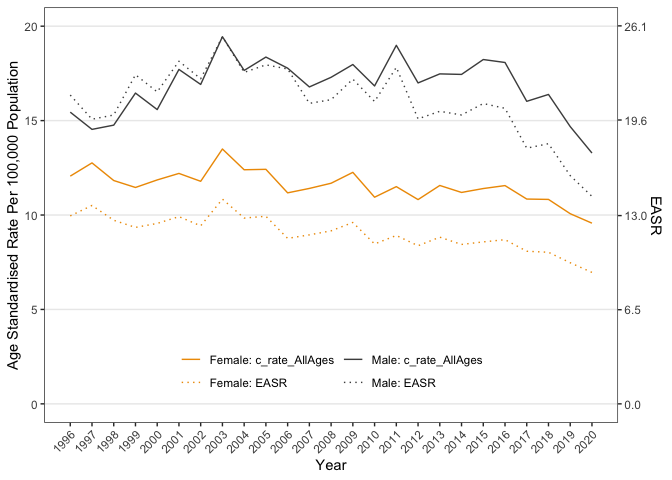
Created on 2023-02-03 by the reprex package (v2.0.1)
My answer to this question explains more about double y-axes in ggplot2.