I have a spreadsheet with Lat-Lon info of 14 regions in the Czech Republic (file 
The sixth column of the spreadsheet has the active cases numbers. I am trying to get the numbers appear as bubbles on the above map. I tried the following but all dots are of the same size. How do I merge plot 1 and plot 2?
my_df <- read.csv("CZE_InitialSeedData.csv", header = T)
class(my_df)
my_sf <- st_as_sf(my_df, coords = c('Lon', 'Lat'))
my_sf <- st_set_crs(my_sf, value = 4326)
my_sf
seedPlot <- ggplot(my_sf)
geom_sf(aes(fill = InitialInfections))
seedPlot <- seedPlot
scale_fill_continuous(name = "Active Cases", low = "pink", high = "red", na.value = "grey50")
seedPlot <- seedPlot
theme(legend.position = "bottom", legend.text.align = 1, legend.title.align = 0.5)
seedPlot
CodePudding user response:
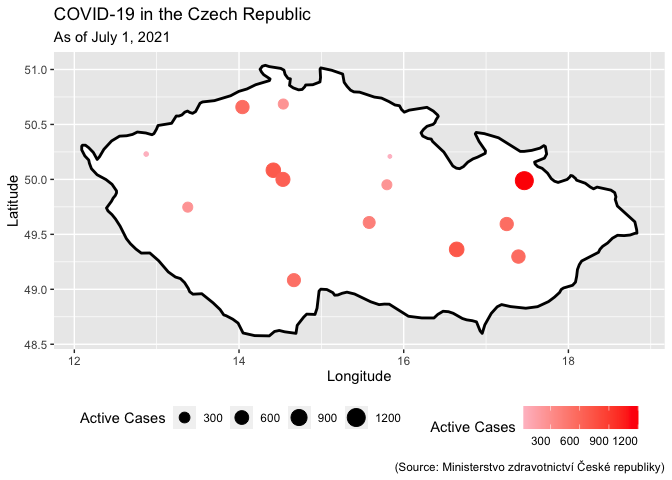
There is no need to convert your data to a sf object. You could simply add your data to your map via a geom_point. To get bubbles map your column with the active cases on the size aesthetic:
library(ggplot2)
library(maps)
library(dplyr)
worldmap <- map_data("world")
worldmap2 <- dplyr::filter(worldmap, region == "Czech Republic")
base_map <- ggplot(worldmap2)
geom_polygon(aes(long, lat, group = group), col = "black", fill = "white", size = 1)
labs(
title = "COVID-19 in the Czech Republic", subtitle = "As of July 1, 2021", x = "Longitude", y = "Latitude",
caption = "(Source: Ministerstvo zdravotnictví České republiky)"
)
base_map
geom_point(
data = my_df,
aes(x = Lon, y = Lat, color = InitialInfections, size = InitialInfections)
)
scale_color_continuous(name = "Active Cases", low = "pink", high = "red", na.value = "grey50")
scale_size_continuous(name = "Active Cases")
theme(legend.position = "bottom", legend.text.align = 1, legend.title.align = 0.5)
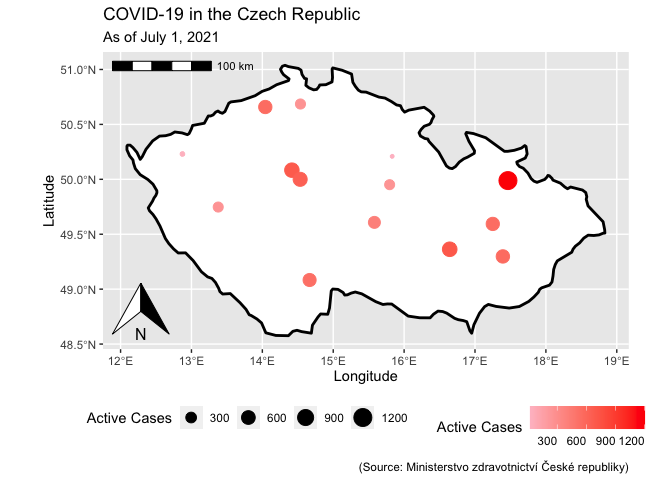
EDIT As far as I get it you could add a north arrow and scale bar for non-sf coords. However, converting to an sf object will automatically pick the right units for the scale bar. To this end convert both the basemap and the points layer to an sf object like so:
library(ggplot2)
library(maps)
library(dplyr)
library(ggspatial)
library(sf)
worldmap <- map_data("world")
worldmap2 <- dplyr::filter(worldmap, region == "Czech Republic") %>%
st_as_sf(coords = c("long", "lat"), crs = 4326) %>%
st_combine() %>%
st_cast("POLYGON")
base_map <- ggplot(worldmap2)
geom_sf(col = "black", fill = "white", size = 1)
annotation_north_arrow()
annotation_scale(location = "tl")
labs(
title = "COVID-19 in the Czech Republic", subtitle = "As of July 1, 2021", x = "Longitude", y = "Latitude",
caption = "(Source: Ministerstvo zdravotnictví České republiky)"
)
my_df <- my_df %>%
st_as_sf(coords = c("Lon", "Lat"), crs = 4326)
base_map
geom_sf(data = my_df, aes(color = InitialInfections, size = InitialInfections))
scale_color_continuous(name = "Active Cases", low = "pink", high = "red", na.value = "grey50")
scale_size_continuous(name = "Active Cases")
theme(legend.position = "bottom", legend.text.align = 1, legend.title.align = 0.5)
DATA
my_df <- structure(list(Location = c(
"Prague", "CentralBohemian", "SouthBohemian",
"Plzen", "KarlovyVary", "UstinadLabem", "Liberec", "HradecKralove",
"Pardubice", "Vysocina", "SouthMoravian", "Olomouc", "Zlin",
"Moravian-Silesian"
), Lat = c(
50.083333, 50, 49.083333, 49.7475,
50.230556, 50.658333, 50.685584, 50.209167, 49.951136, 49.6079,
49.363161, 49.593889, 49.29786, 49.988449
), Lon = c(
14.416667,
14.533333, 14.666667, 13.3775, 12.8725, 14.041667, 14.537747,
15.831944, 15.795636, 15.580728, 16.643175, 17.250833, 17.393135,
17.464759
), InitialVaccinated = c(
252944L, 159560L, 93490L, 82014L,
40129L, 104454L, 59442L, 82074L, 65060L, 66325L, 165250L, 89116L,
80125L, 159490L
), InitialExposed = c(
1380L, 1274L, 1048L, 500L,
50L, 1098L, 506L, 42L, 492L, 820L, 1406L, 1090L, 1116L, 2404L
), InitialInfections = c(
690L, 637L, 524L, 250L, 25L, 549L, 253L,
21L, 246L, 410L, 703L, 545L, 558L, 1202L
), InitialRecovered = c(
181947L,
226944L, 97405L, 95944L, 43882L, 120416L, 79029L, 102835L, 91729L,
78308L, 151627L, 90887L, 89163L, 174251L
), InitialDead = c(
2736L,
3421L, 1978L, 1912L, 1484L, 2523L, 1280L, 1811L, 1437L, 1375L,
3412L, 1709L, 1594L, 3521L
)), class = "data.frame", row.names = c(
NA,
-14L
))