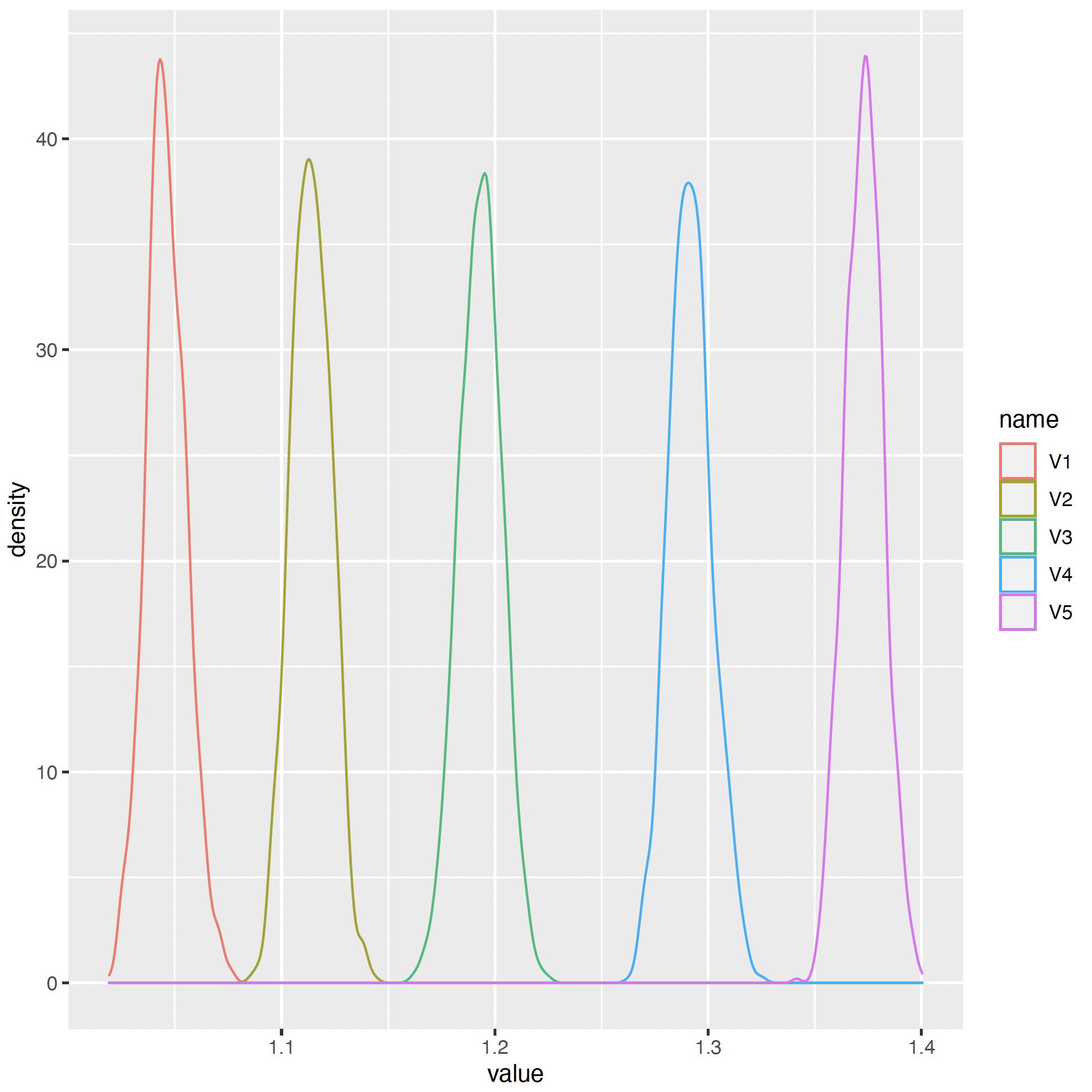
Below the result of a R script :
this R code snippet is :
as.data.frame(y3) %>%
mutate(row = row_number()) %>% # add row to simplify next step
pivot_longer(-row) %>% # reshape long
ggplot(aes(value, color = name)) # map x to value, color to name
geom_density()
how can I change the name of xlabel (value) and ylabel (density) and the legend also (v1, v2, v3, v4, v5) ?
UPDATE 1:
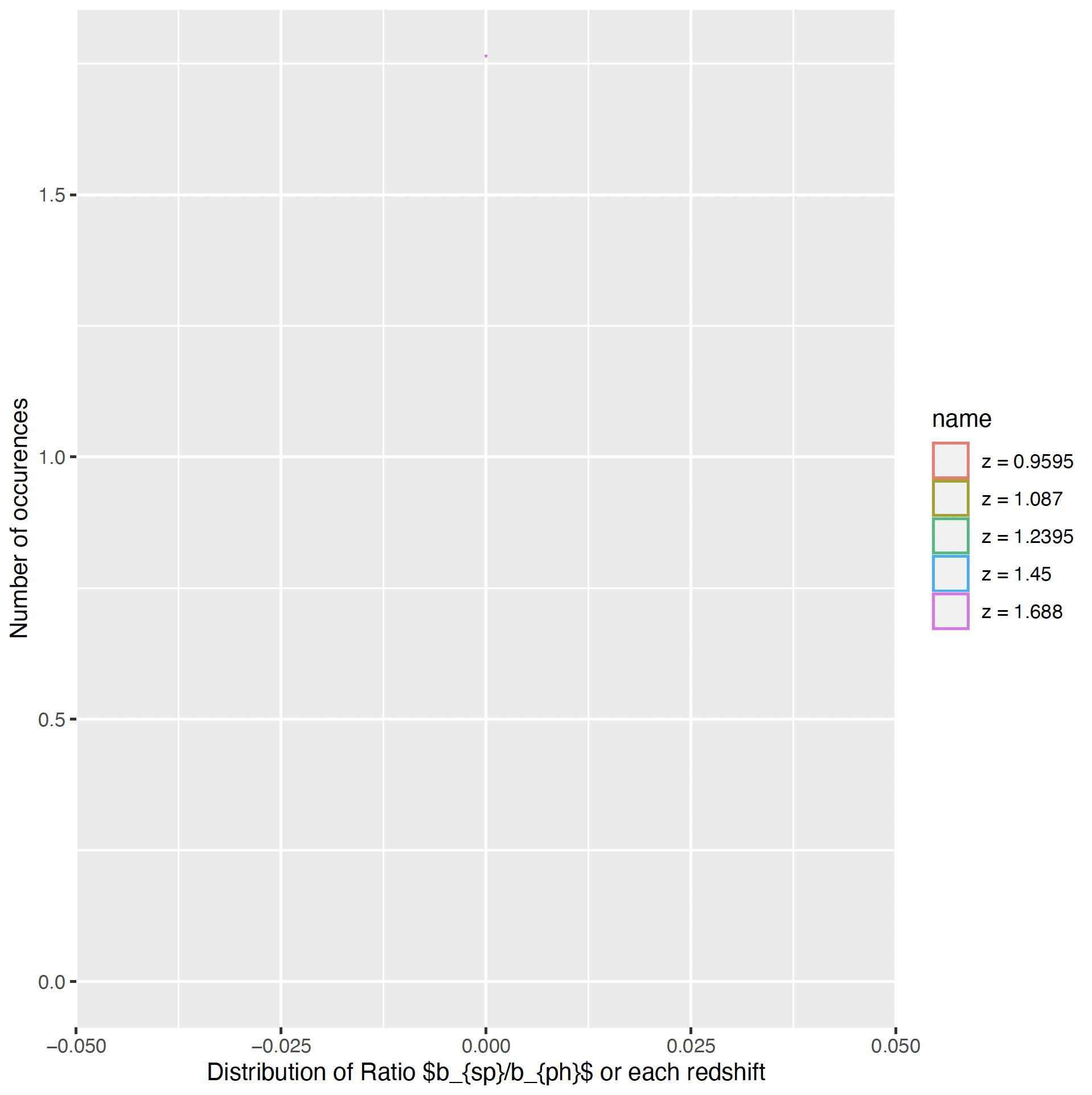
By using the code snippet of @Park, I get no curves plotted :
as.data.frame(y3) %>%
mutate(row = row_number()) %>% # add row to simplify next step
pivot_longer(-row) %>% # reshape long
mutate(name = recode(name, V1="z = 0.9595", V2="z = 1.087", V3="z = 1.2395", V4="z = 1.45", V5="z = 1.688")) %>%
ggplot(aes(value, color = name)) # map x to value, color to name
geom_density()
xlab("Distribution of Ratio $b_{sp}/b_{ph}$ or each redshift")
ylab("Number of occurences")
and the result :
I tried also to use subscript with Latex format : $b_{sp}/b_{ph}$ but without success.
CodePudding user response:
You may try xlab, ylab, scale_color_manual,
as.data.frame(y3) %>%
mutate(row = row_number()) %>% # add row to simplify next step
pivot_longer(-row) %>% # reshape long
ggplot(aes(value, color = name)) # map x to value, color to name
geom_density()
xlab("text")
ylab("text")
scale_color_manual(labels = c("a", "b", "c", "d", "e"))
Recode before plot
as.data.frame(y3) %>%
mutate(row = row_number()) %>% # add row to simplify next step
pivot_longer(-row) %>% # reshape long
mutate(name = recode(name, V1 = "a", V2 = "b", V3 = "c", V4 = "d", V5 = "e")) %>%
ggplot(aes(value, color = name)) # map x to value, color to name
geom_density()
xlab("text")
ylab("text")
Using Array_total_WITH_Shot_Noise data
my_data <- read.delim("D:/Prac/Array_total_WITH_Shot_Noise.txt", header = FALSE, sep = " ")
array_2D <- array(my_data)
z_ph <- c(0.9595, 1.087, 1.2395, 1.45, 1.688)
b_sp <- c(1.42904922, 1.52601862, 1.63866958, 1.78259615, 1.91956918)
b_ph <- c(sqrt(1 z_ph))
ratio_squared <- (b_sp/b_ph)^2
nRed <- 5
nRow <- NROW(my_data)
nSample_var <- 1000000
nSample_mc <- 1000
Cl<-my_data[,2:length(my_data)]#suppose cl=var(alm)
Cl_sp <- array(0, dim=c(nRow,nRed))
Cl_ph <- array(0, dim=c(nRow,nRed))
length(Cl)
for (i in 1:length(Cl)) {
#(shape/rate) convention :
Cl_sp[,i] <-(Cl[, i] * ratio_squared[i])
Cl_ph[,i] <- (Cl[, i])
}
L <- array_2D[,1]
L <- 2*(array_2D[,1]) 1
# Weighted sum of Chi squared distribution
y3_1<-array(0,dim=c(nSample_var,nRed));y3_2<-array(0,dim=c(nSample_var,nRed));y3<-array(0,dim=c(nSample_var,nRed));
for (i in 1:nRed) {
for (j in 1:nRow) {
# Try to summing all the random variable
y3_1[,i] <- y3_1[,i] Cl_sp[j,i] * rchisq(nSample_var,df=L[j])
y3_2[,i] <- y3_2[,i] Cl_ph[j,i] * rchisq(nSample_var,df=L[j])
}
y3[,i] <- y3_1[,i]/y3_2[,i]
}
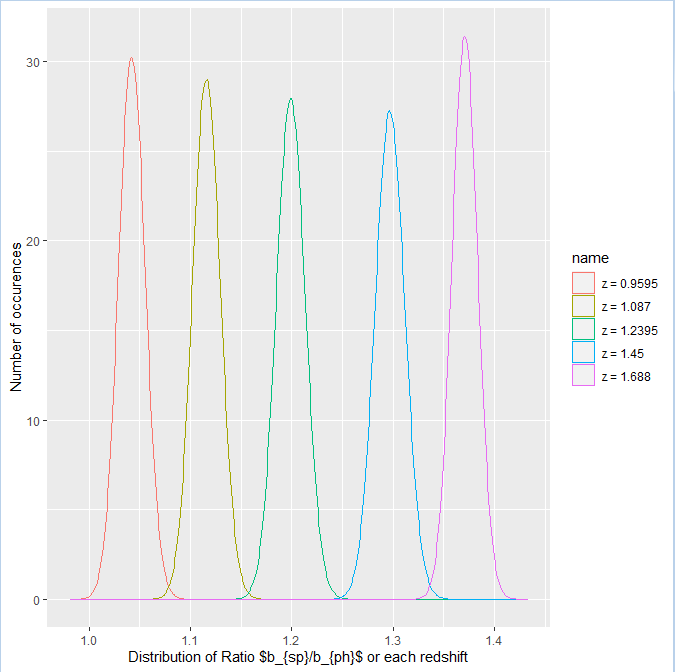
as.data.frame(y3) %>%
mutate(row = row_number()) %>% # add row to simplify next step
pivot_longer(-row) %>% # reshape long
mutate(name = recode(name, V1="z = 0.9595", V2="z = 1.087", V3="z = 1.2395", V4="z = 1.45", V5="z = 1.688")) %>%
ggplot(aes(value, color = name)) # map x to value, color to name
geom_density()
xlab(TeX("Distribution of Ratio $b_{sp}/b_{ph}$ or each redshift"))
ylab("Number of occurences")