I'm trying to make my ODEs solver faster with Numba, but the following code throws a Typing Error:
import numpy as np
import matplotlib.pyplot as plt
from numba import njit
@njit
def pend(t, y, b, c):
theta, omega = y
dydt = np.array([omega, -b*omega - c*np.sin(theta)])
return dydt
@njit
def rungeStep(f, t, y0, tau, params):
k1 = tau * f(t, y0, *params)
k2 = tau * f(t, y0 k1 / 2, *params)
k3 = tau * f(t, y0 k2 / 2, *params)
k4 = tau * f(t, y0 k3, *params)
return (k1 2 * k2 2 * k3 k4) / 6
@njit
def integrate(f, t0, y0, tEnd, h, params):
ys = y0.copy()
t = np.array(t0)
while t0 <= tEnd:
y0 = rungeStep(f, t0, y0[0], h, params)
t0 = h
ys = np.concatenate((ys, y0), axis=0)
t = np.append(t, t0)
return t, ys.T
args = (0.25, 5)
y0 = np.array([[np.pi - 0.1, 0.0]])
t, y = integrate(pend, 0, y0, 10, 1, args)
This results in:
TypingError: Failed in nopython mode pipeline (step: nopython frontend)
Cannot unify array(int64, 0d, C) and array(int64, 1d, C) for 't.2', defined at <ipython-input-56-38d2ea70b889> (6)
File "<ipython-input-56-38d2ea70b889>", line 6:
def inagrate(f, t0, y0, tEnd, h, params):
<source elided>
while t0 <= tEnd:
y0 = rungeStep(f, t0, y0[0], h, params)
^
During: typing of assignment at <ipython-input-56-38d2ea70b889> (6)
File "<ipython-input-56-38d2ea70b889>", line 6:
def inagrate(f, t0, y0, tEnd, h, params):
<source elided>
while t0 <= tEnd:
y0 = rungeStep(f, t0, y0[0], h, params)
^
Without the njit-decorator it works fine. Can anybody help me please?
CodePudding user response:
One should get the same error without JIT, but perhaps the broadcasting rules are flexible enough. Your y0 is a 2d array, what you pass to rungeStep (of the Heun-Kutta method) is a 1d array, as is its return value. So you can not use that result immediately to update y0, you would have to update y0[0]. But then the question is why introduce this complexity in the first place.
Numpy arrays are optimized as fixed-size objects, appending to them requires memory allocation and copying in every instance, which is slower than appending to lists, where re-allocation is done with a (growing) reserve. Numba-JIT appears to have a problem with transforming lists of numpy arrays to 2d numpy arrays. What works is to manually downgrade the arrays to simple lists. With some trial-and-error I got the following code running (only repeating pieces with changes):
@njit
def rungeStep(f, t, y0, tau, params):
k1 = tau * f(t, y0, *params)
k2 = tau * f(t tau/2, y0 k1 / 2, *params)
k3 = tau * f(t tau/2, y0 k2 / 2, *params)
k4 = tau * f(t tau, y0 k3, *params)
return (k1 2 * k2 2 * k3 k4) / 6
@njit
def integrate(f, t0, y0, tEnd, h, params):
ys = [list(y0)]
t = [t0]
while t0 <= tEnd:
y0 = rungeStep(f, t0, y0, h, params)
t0 = h
ys.append(list(y0))
t.append(t0)
return np.array(t), np.array(ys).T
args = (0.25, 5.0)
y0 = np.array([np.pi - 0.1, 0.0])
t, y = integrate(pend, 0, y0, 10, 1e-1, args)
Note that your RK4 implementation had some omissions, not critical here but an error source for time-depending ODE examples.
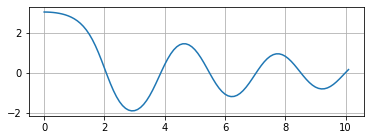
Plotting the result gives the reasonable graph