I am trying to skip NA values on a graph with two lines on it (ad13com & Pd13c), and combining it with a second graph (xPDd13C) using grid arrange. Using my code, the line breaks due to NA values in my data and I am looking for continuous lines (three separate ones). I have tried using na.rm arguments but didn't make any progress as I'm quite new to r. I realise that the issue is coming from the fact that I am plotting the ungathered data but I don't know how to apply a skip NA argument to data thats being plotted this way!
In my code, I first gather the data before plotting it as I am using a similar script for when I produced multiple plots using facet wrap, so it may not be necessary to gather the data for this application? I also realise I have multiple y axis scales so it just replaces the existing scale and uses the last one, I wasn't too sure how to remove it properly so I just left it in as it works fine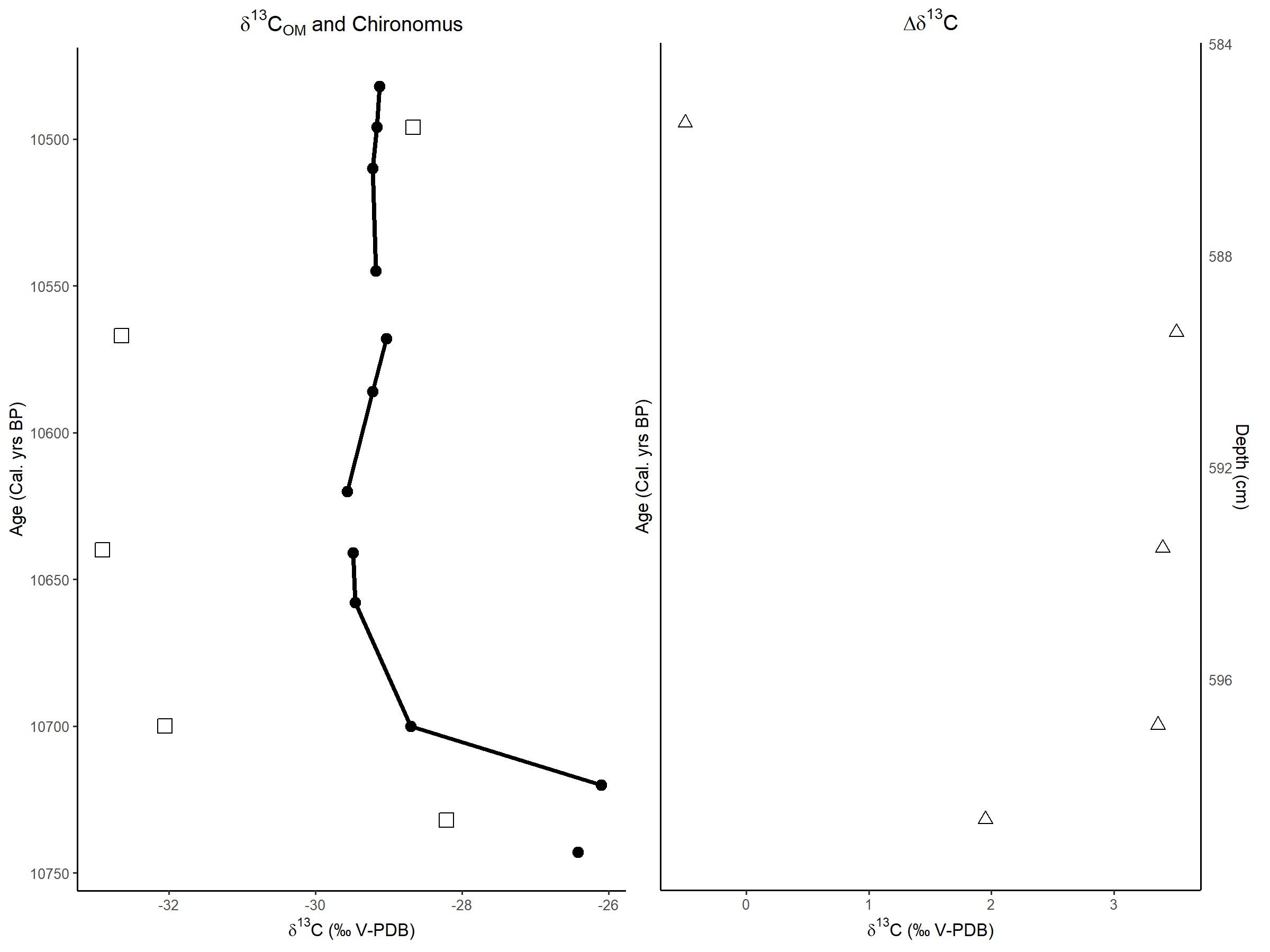
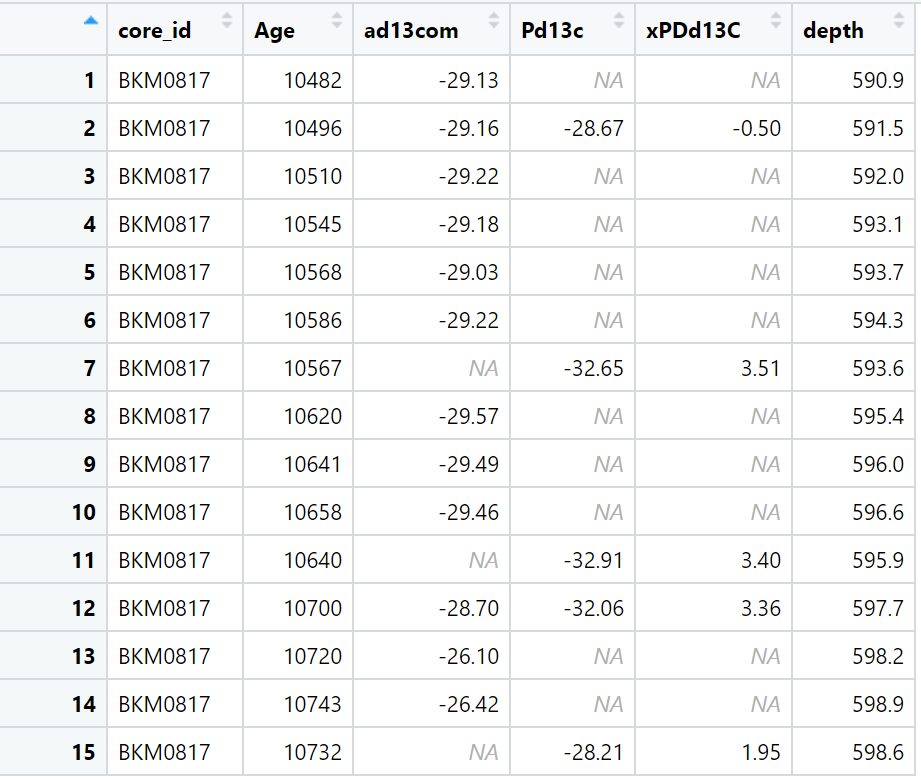
Anyway here is my code:
theme_set(theme_paleo(8))
theme_update(plot.title = element_text(hjust = 0.5))
#read the data
data <- read_csv("profundal.csv", col_types = cols(.default = col_guess())
)
#first gather the data and set out the omit na function in order to skip na values
data %>% filter(core_id == "BKM0817") %>%
gather(key = param, value = value, -core_id, -Age, -depth) %>%
na.omit()
#plot the first graph
prof1 <-
ggplot()
geom_lineh(data = data, mapping = aes(x=ad13com, y = Age), colour = "black", size = 1)
geom_point(data = data, mapping = aes(x=ad13com, y = Age), colour = "black", size = 2)
geom_lineh(data = data, mapping = aes(x=Pd13c, y = Age), linetype = 2, colour = "black", size = 1,)
geom_point(data = data, mapping = aes(x=Pd13c, y = Age), shape=0, colour = "black", size = 2.7)
scale_y_reverse()
labs (x = expression(delta ^13*"C (\u2030 V-PDB)"), y = "Age (Cal. yrs BP)")
ggtitle(expression(delta ^13*"C"[OM]~"and Chironomus"))
theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black"))
#plot the second graph
prof2 <-
ggplot()
geom_lineh(data = data, mapping = aes(x=xPDd13C, y = Age), colour = "black", size = 1)
geom_point(data = data, mapping = aes(x=xPDd13C, y = Age), shape = 2, colour = "black", size = 2)
scale_y_reverse(# Features of the first axis
name = "Age (Cal. yrs BP)",
# Add a second axis and specify its features
sec.axis = sec_axis( trans=~./17.927, name="Depth (cm)")
)
labs(x = expression(delta ^13*"C (\u2030 V-PDB)"), y = "Age (Cal. yrs BP)")
ggtitle( expression(Delta*delta ^13*"C"))
theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black"))
#combine the plots
profundal <-gridExtra::grid.arrange(
prof1,
prof2
labs(y = NULL)
scale_y_reverse(labels = NULL, name = "Age (Cal. yrs BP)",
# Add a second axis and specify its features
sec.axis = sec_axis( trans=~./17.927, name="Depth (cm)"))
theme(
plot.margin = unit(c(0.05,0.1, 0.056,0), "inches"),
axis.ticks.y = element_blank(),
plot.background = element_blank()
),
nrow = 1,
widths = c(4, 4)
)
# save the file
ggsave("test.png", units="in", width=8, height=6, dpi=300, plot=profundal)
Many thanks in advance!
CodePudding user response:
Where you have used na_omit() on data the results are not assigned to anything. You need to assign the results to data (or something else).
data <- data %>% filter(core_id == "BKM0817") %>%
gather(key = param, value = value, -core_id, -Age, -depth) %>%
na.omit()
CodePudding user response:
I managed to plot the new data after it was gathered (as Tjn suggested with the na.omit function at the end) like this.
data2 <- data %>% filter(core_id == "BKM0817") %>%
gather(key = param, value = value, -core_id, -Age, -depth) %>%
na.omit()
The data then looks like this: 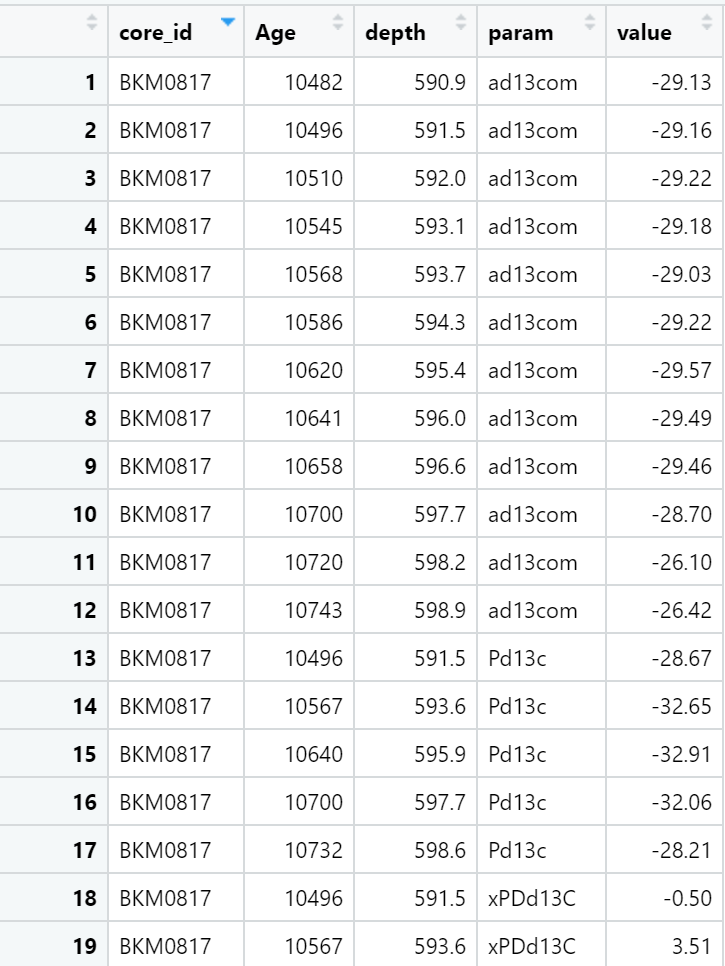
In order to plot two lines on the same graph, I then had to subset the data and plot the lines and points individually like this for the first graph and follows the same structure for the second graph:
prof1 <-
ggplot()
geom_lineh(data = subset(data2,param %in% "ad13com"), mapping = aes(x=value, y = Age), colour = "black", size = 1)
geom_point(data = subset(data2,param %in%"ad13com"), mapping = aes(x=value, y = Age), colour = "black", size = 2)
geom_lineh(data = subset(data2,param %in% "Pd13c"), mapping = aes(x=value, y = Age), linetype = 2, colour = "black", size = 1,)
geom_point(data = subset(data2,param %in% "Pd13c"), mapping = aes(x=value, y = Age), shape=0, colour = "black", size = 2.7)
scale_y_reverse()
labs (x = expression(delta ^13*"C (\u2030 V-PDB)"), y = "Age (Cal. yrs BP)")
ggtitle(expression(delta ^13*"C"[OM]~"and Chironomus"))
theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black"))
The final result is now the same as the graph in the question but now with lines linking all the points individually.
