I have this graph structure
graph = thisdict['data']['graph']
dict_edges = graph['edges']
edges = []
for edge in dict_edges:
edges.append((edge['source']['node_id'], edge['target']['node_id']))
print('Source:\t\t\t Target:\n')
for edge in edges:
print(str(edge))
print('\n')
dict_nodes = graph['nodes']
nodes = {}
for node in dict_nodes:
nodes[node['id']] = node['name']
print('Node ID:\t\t Node Name:\n')
for key, value in nodes.items():
print("'%s':'%s'" %(key, value))
The output:
Source: Target:
('61697b94f74c92a808641ba3', '61697b95f74c92a808641ba4')
('61697b94f74c92a808641ba3', '61697b96f74c92a808641ba5')
('61697b95f74c92a808641ba4', '61697b96f74c92a808641ba6')
('61697b96f74c92a808641ba6', '61697b97f74c92a808641ba7')
('61697b96f74c92a808641ba5', '61697b97f74c92a808641ba7')
('61697b97f74c92a808641ba7', '61697b98f74c92a808641ba8')
('61697b98f74c92a808641ba8', '61697b98f74c92a808641ba9')
Node ID: Node Name:
'61697b94f74c92a808641ba3':'S3 connector'
'61697b95f74c92a808641ba4':'loader 1'
'61697b96f74c92a808641ba5':'loader 2'
'61697b96f74c92a808641ba6':'sampler 1'
'61697b97f74c92a808641ba7':'concator'
'61697b98f74c92a808641ba8':'sampler 2'
'61697b98f74c92a808641ba9':'splitter'
I wrote this code to draw the graph:
nx_graph = nx.Graph()
plt.figure(figsize=(3,3))
for key, value in nodes.items():
nx_graph.add_node(key, layer = nodes.values())
#I need to put every node name in a single layer, So I should have 6 layers
for edge in edges:
nx_graph.add_edge(*edge)
pos = nx.multipartite_layout(nx_graph, subset_key="layer")
nx.draw(nx_graph, pos, labels=nodes, with_labels=True)
plt.show()
It shows an error says : TypeError: unsupported operand type(s) for -: 'dict_values' and 'float'
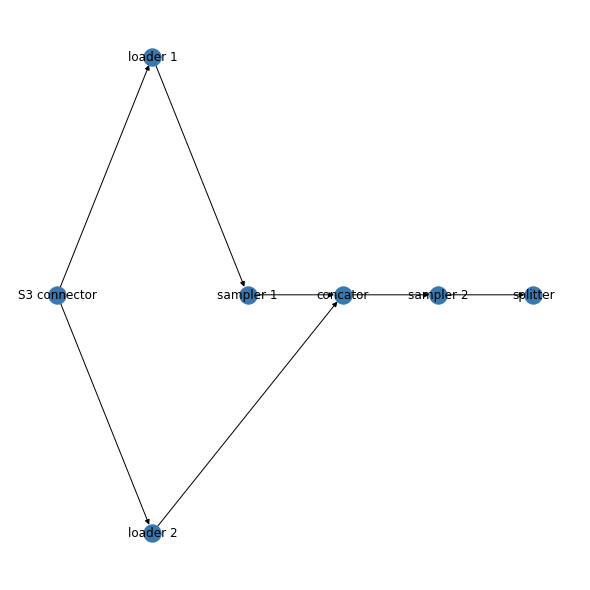
I need to put every node name in a single layer, So I should have 6 layers. The layers sorting should be like the following:
S3 connector --> loader 1 & loader 2
loader 1 will give sampler 1
loader 2 & sampler 1 will meet in concator
concator will give sampler 2
sampler 2 will give splitter
CodePudding user response:
The error is occurring because when you add nodes in the for loop, you pass layer = nodes.values() instead of just layer=value. However, correcting that still won't give you the layout that you want because you actually have to specify the layers somehow. Based on your description of the 6 layers, I added them as attributes from a separate dictionary.
I made some assumptions about what you're actually trying to do by changing to a DiGraph and deciding where the sampler 1 nod is.
Here is a self-contained code block and its output.
import networkx as nx
import matplotlib.pyplot as plt
nodes = {'61697b94f74c92a808641ba3':'S3 connector',
'61697b95f74c92a808641ba4':'loader 1',
'61697b96f74c92a808641ba5':'loader 2',
'61697b96f74c92a808641ba6':'sampler 1',
'61697b97f74c92a808641ba7':'concator',
'61697b98f74c92a808641ba8':'sampler 2',
'61697b98f74c92a808641ba9':'splitter'}
edges = [('61697b94f74c92a808641ba3', '61697b95f74c92a808641ba4'),
('61697b94f74c92a808641ba3', '61697b96f74c92a808641ba5'),
('61697b95f74c92a808641ba4', '61697b96f74c92a808641ba6'),
('61697b96f74c92a808641ba6', '61697b97f74c92a808641ba7'),
('61697b96f74c92a808641ba5', '61697b97f74c92a808641ba7'),
('61697b97f74c92a808641ba7', '61697b98f74c92a808641ba8'),
('61697b98f74c92a808641ba8', '61697b98f74c92a808641ba9')]
layers = {'61697b94f74c92a808641ba3': 1,
'61697b95f74c92a808641ba4': 2,
'61697b96f74c92a808641ba5':2,
'61697b96f74c92a808641ba6':3,
'61697b97f74c92a808641ba7':4,
'61697b98f74c92a808641ba8':5,
'61697b98f74c92a808641ba9':6}
nx_graph = nx.DiGraph() # Made this a DiGraph (adds arrows to visual)
plt.figure(figsize=(8,8)) # Enlarged figure
for key, value in nodes.items():
nx_graph.add_node(key, name=value, layer=layers[key])
for edge in edges:
nx_graph.add_edge(*edge)
pos = nx.multipartite_layout(nx_graph, subset_key="layer")
nx.draw(nx_graph, pos=pos, labels=nodes, with_labels=True)
plt.show()