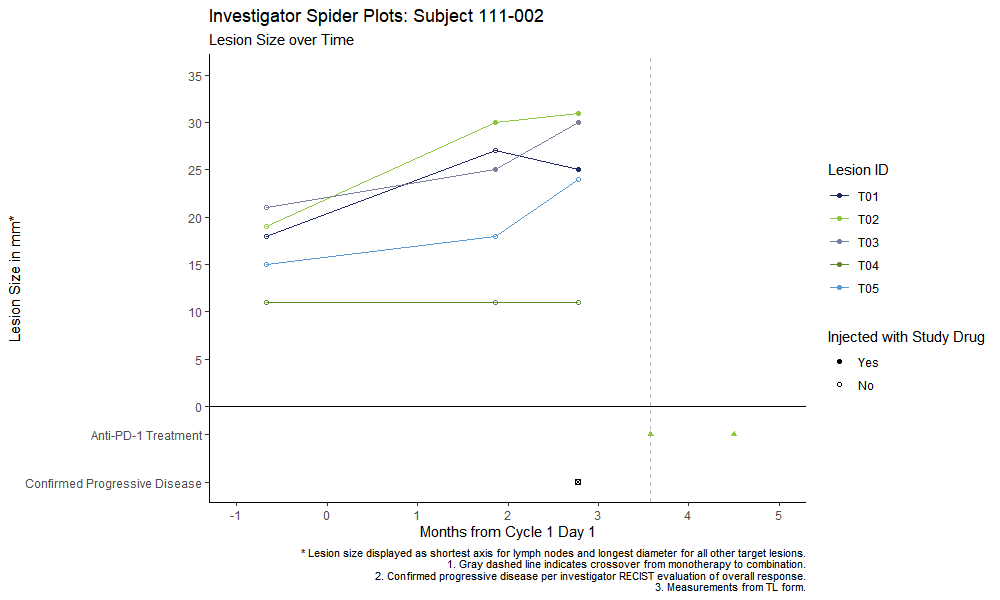
I have some complex ggplot code for some spider plots. I was told that instead of including the information in the caption that folks want to see it as part of the legend. However, a lot of the items are not part of a particular scale. Specifically folks want to see the dashed gray line as a horizontal gray dashed line to signify crossover, the green triangle to signify anti-PD-1 dosing, and the box with the x in the middle (shape number 7) to signify progressive disease.
Would it be possible to draw these elements directly onto the plot? My initial inclination was to use geom_rect and geom_text to draw directly onto the plot, but I figured I would see if it is possible to add it within the legend first.
Here is an example plot, with some identifying info removed:
Here is the corresponding code:
pallete <- c("#202960", "#8CC63E", "#797e9f", "#628a2b", "#5B9BD5", "#8f94af", "#bebebe", "#a5a9bf", "#46631f")
example <- ggplot()
#scatter plot of target lesion
geom_point(data = target_data,
#if lymph node flag present, plot short axis instead of long diam
aes(x = mos_dur, y = if_else(lymphnode_flag == 1, short_axis, long_diam),
group = les_id, colour = les_id,
shape = trt_flag), na.rm = TRUE)
#connect the lines, grouping by target lesion
geom_line(data = target_data,
aes(x = mos_dur, y = if_else(lymphnode_flag == 1, short_axis, long_diam),
group = les_id, colour = les_id), na.rm = TRUE)
theme_classic()
labs(title = paste("Investigator Spider Plots: Subject 111-002"),
subtitle = "Lesion Size over Time",
caption = paste0("* Lesion size displayed as shortest axis for lymph nodes and longest diameter for all other target lesions.\n1. Gray dashed line indicates crossover from monotherapy to combination.\n2. Confirmed progressive disease per investigator RECIST evaluation of overall response.\n3. Measurements from TL form."),
color = "Lesion ID",
shape = "Injected with Study Drug")
geom_hline(aes(yintercept = 0), color = "black")
geom_vline(data = cross_data, aes(xintercept = mos_dur), color = "gray", linetype = "dashed")
geom_point(data = prog_data, aes(x = mos_dur, y = -8), shape = 7)
geom_point(data = pd1_data,
aes(x = mos_dur, y = -3), color = pallete[2], shape = 17)
scale_x_continuous("Months from Cycle 1 Day 1", breaks = c(-1,0,1,2,3,4,5))
scale_y_continuous("Lesion Size in mm*", breaks = c(-8, -3, seq(0,70,5)),
labels = c("Confirmed Progressive Disease", "Anti-PD-1 Treatment",
as.character(seq(0,70,5))))
scale_color_manual(values = pallete[1:5]) #, breaks = c("T01", "T02", "T03", "T04"))
scale_shape_manual(values = c(16,1), breaks = c(1,0), labels = c("Yes", "No"))
guides(color = guide_legend(order = 1), shape = guide_legend(order = 2))
coord_cartesian(xlim = c(-1,5), ylim = c(-8,35))
theme(plot.caption = element_text(size = 8))
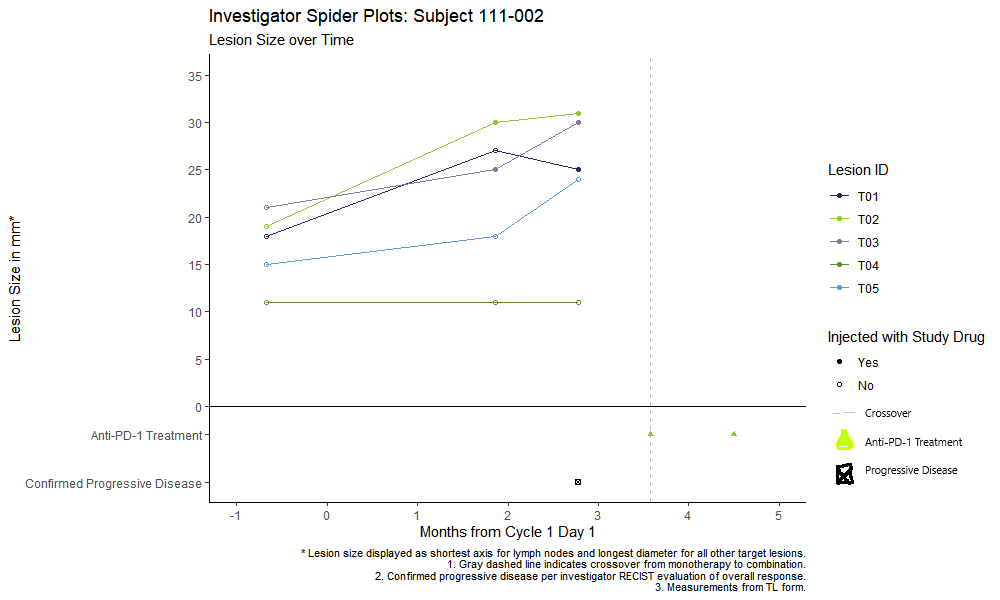
Here is a quick and dirty mockup of what folks want to see (excuse the fact that it was done in MS paint):
regex of datasets:
target_data <- structure(list(subjid = c("111-002", "111-002", "111-002", "111-002",
"111-002", "111-002", "111-002", "111-002", "111-002", "111-002",
"111-002", "111-002", "111-002", "111-002", "111-002"), day1_dat = structure(c(1621468800,
1621468800, 1621468800, 1621468800, 1621468800, 1621468800, 1621468800,
1621468800, 1621468800, 1621468800, 1621468800, 1621468800, 1621468800,
1621468800, 1621468800), tzone = "UTC", class = c("POSIXct",
"POSIXt")), visit = c("Screening", "Screening", "Screening",
"Screening", "Screening", "Treatment Visit Cycle", "Treatment Visit Cycle",
"Treatment Visit Cycle", "Treatment Visit Cycle", "Treatment Visit Cycle",
"Treatment Visit Cycle", "Screening", "Treatment Visit Cycle",
"Treatment Visit Cycle", "Treatment Visit Cycle"), visdat = structure(c(18747,
18747, 18747, 18747, 18747, 18824, 18824, 18824, 18824, 18824,
18852, 18852, 18852, 18852, 18852), class = "Date"), mos_dur = c(-0.666666666666667,
-0.666666666666667, -0.666666666666667, -0.666666666666667, -0.666666666666667,
1.86666666666667, 1.86666666666667, 1.86666666666667, 1.86666666666667,
1.86666666666667, 2.7741935483871, 2.7741935483871, 2.7741935483871,
2.7741935483871, 2.7741935483871), cycle = c(NA, NA, NA, NA,
NA, 3, 3, 3, 3, 3, 4, NA, 4, 4, 4), les_flag = c("Target", "Target",
"Target", "Target", "Target", "Target", "Target", "Target", "Target",
"Target", "Target", "Target", "Target", "Target", "Target"),
les_id = c("T01", "T02", "T04", "T05", "T03", "T02", "T03",
"T04", "T01", "T05", "T03", "T04", "T02", "T05", "T01"),
les_site = c("LYMPH NODE", "SKIN", "LUNG", "SKIN", "SKIN",
"SKIN", "SKIN", "LUNG", "LYMPH NODE", "SKIN", "SKIN", "LUNG",
"SKIN", "SKIN", "LYMPH NODE"), lymphnode_flag = structure(c(2L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 2L), .Label = c("0",
"1"), class = "factor"), other_spec = c("", "", "", "", "",
"", "", "", "", "", "", "", "", "", ""), les_details = c("right axilla",
"left back", "left upper lobe", "right thigh", "right back",
"left back", "right back", "left upper lobe", "right axilla",
"right thigh", "right back", "left upper lobe", "left back",
"right thigh", "right axilla"), long_diam = c(21, 19, 11,
15, 21, 30, 25, 11, 30, 18, 30, 11, 31, 24, 31), short_axis = c(18,
11, 11, 7, 11, 20, 18, 10, 27, 11, 19, 10, 26, 12, 25), les_stat = c("Present, Measured",
"Present, Measured", "Present, Measured", "Present, Measured",
"Present, Measured", "Present, Measured", "Present, Measured",
"Present, Measured", "Present, Measured", "Present, Measured",
"Present, Measured", "Present, Measured", "Present, Measured",
"Present, Measured", "Present, Measured"), prog_flag = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L), .Label = c("0",
"1"), class = c("ordered", "factor")), trt_flag = structure(c(1L,
1L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 2L), .Label = c("0",
"1"), class = "factor")), row.names = c(NA, -15L), groups = structure(list(
subjid = c("111-002", "111-002"), visit = c("Screening",
"Treatment Visit Cycle"), .rows = structure(list(c(1L, 2L,
3L, 4L, 5L, 12L), c(6L, 7L, 8L, 9L, 10L, 11L, 13L, 14L, 15L
)), ptype = integer(0), class = c("vctrs_list_of", "vctrs_vctr",
"list"))), row.names = c(NA, -2L), class = c("tbl_df", "tbl",
"data.frame"), .drop = TRUE), class = c("grouped_df", "tbl_df",
"tbl", "data.frame"))
prog_data <- structure(list(subjid = structure("111-002", label = "Subject name or identifier", format.sas = "$"),
mos_dur = 2.7741935483871), row.names = c(NA, -1L), class = c("tbl_df",
"tbl", "data.frame"))
pd1_data <- structure(list(subjid = structure(c("111-002", "111-002"), label = "Subject name or identifier", format.sas = "$"),
day1_dat = structure(c(18767, 18767), label = "Visit Date", format.sas = "DATETIME", class = "Date"),
admin_dat = structure(c(18877, 18905), label = "Infusion Date", format.sas = "DATETIME", class = "Date"),
mos_dur = c(3.58064516129032, 4.5), visit = structure(c("Crossover Treatment Visit Cycle 5",
"Crossover Treatment Visit Cycle 6"), label = "Folder instance name", format.sas = "$"),
trt = c("Anti-PD-1", "Anti-PD-1"), admin_flag = structure(c("Yes",
"Yes"), label = "Was drug administered at this visit", format.sas = "$"),
interuption_flag = structure(c("No", "No"), label = "Was infusion interrupted at this visit", format.sas = "$"),
intended_flag = structure(c(NA_character_, NA_character_), label = "Was intended volume administered?", format.sas = "$"),
dose = structure(c(480, 480), label = "Actual Dose Administered"),
les_id = c(NA_character_, NA_character_), les_msmt = structure(c(NA_real_,
NA_real_), label = "Lesion Measurement")), row.names = c(NA,
-2L), class = c("tbl_df", "tbl", "data.frame"))
cross_data <- structure(list(subjid = structure("111-002", label = "Subject name or identifier", format.sas = "$"),
crossover_dt = structure(1630972800, tzone = "UTC", class = c("POSIXct",
"POSIXt")), day1_dat = structure(1621468800, label = "Visit Date", tzone = "UTC", format.sas = "DATETIME", class = c("POSIXct",
"POSIXt")), mos_dur = 3.58064516129032), row.names = c(NA,
-1L), class = c("tbl_df", "tbl", "data.frame"))
CodePudding user response:
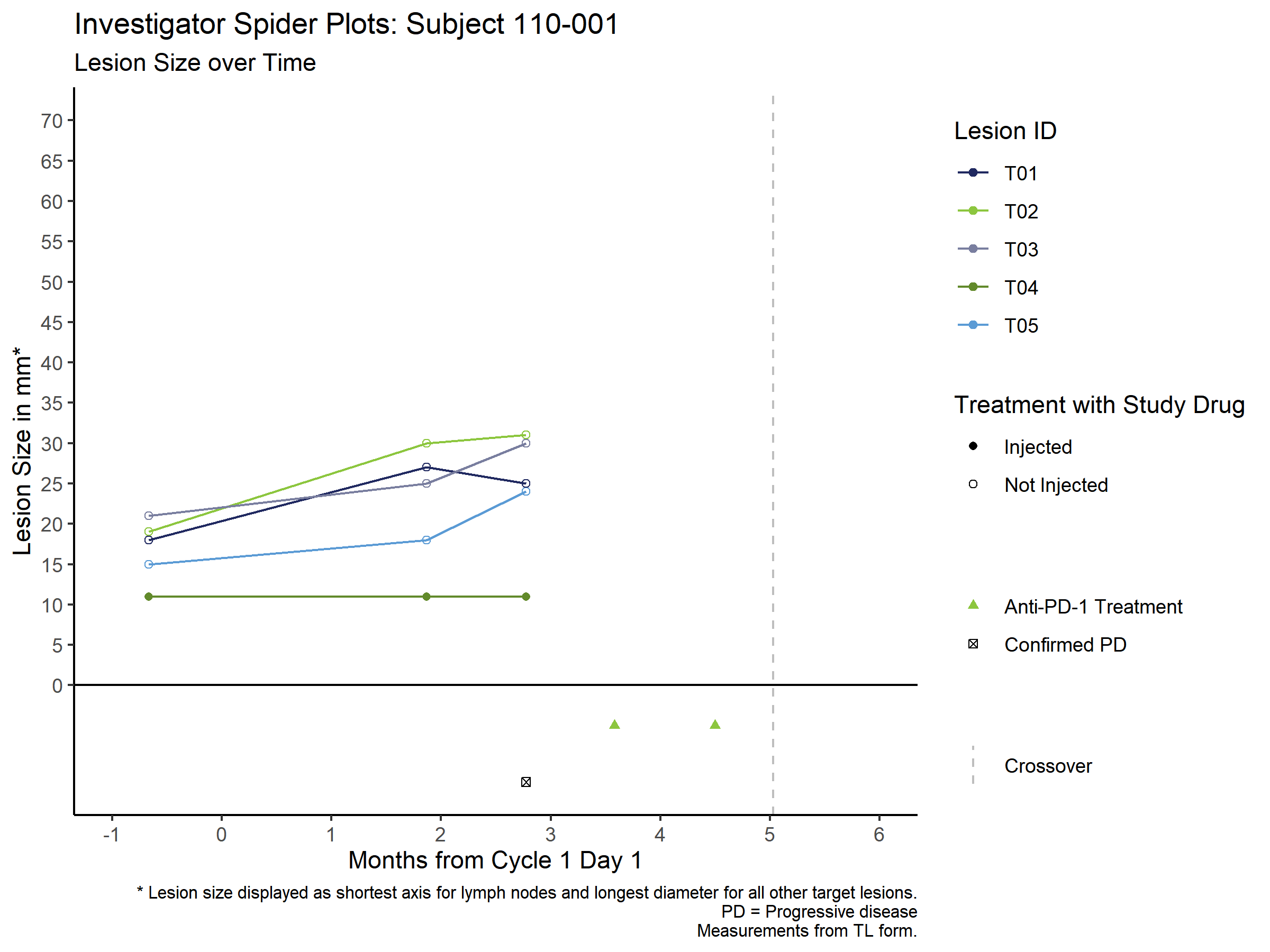
I was able to do some serious "finessing", but I got the desired output.
Solution code and image of output below.
example <- ggplot()
#scatter plot of target lesion
geom_point(data = target_data,
#if lymph node flag present, plot short axis instead of long diam
aes(x = mos_dur, y = if_else(lymphnode_flag == 1, short_axis, long_diam),
group = les_id, colour = les_id,
shape = if_else(trt_flag == 1, "Injected", "Not Injected")), na.rm = TRUE)
#connect the lines, grouping by target lesion
geom_line(data = target_data,
aes(x = mos_dur, y = if_else(lymphnode_flag == 1, short_axis, long_diam),
group = les_id, colour = les_id), na.rm = TRUE)
theme_classic()
labs(title = paste("Investigator Spider Plots: Subject 110-001"),
subtitle = "Lesion Size over Time",
caption = paste0("* Lesion size displayed as shortest axis for lymph nodes and longest diameter for all other target lesions.\nPD = Progressive disease\nMeasurements from TL form."),
color = "Lesion ID",
shape = "Injected with Study Drug")
geom_hline(aes(yintercept = 0), color = "black")
#manually entered crossover - Used date for treatment cycle 9 as placeholder
geom_vline(data = cross_data, aes(xintercept = 5.0322581, linetype = "Crossover"), color = "gray")
geom_point(data = prog_data, aes(x = mos_dur, y = -12, alpha = "PD"), shape = 7)
geom_point(data = pd1_data,
aes(x = mos_dur, y = -5, alpha = "PD-1"), color = palette[2], shape = 17)
scale_x_continuous("Months from Cycle 1 Day 1", breaks = c(seq(-1,6,1)))
scale_y_continuous("Lesion Size in mm*", breaks = c(seq(0,70,5)))
scale_color_manual(values = palette[1:5]) #, breaks = c("T01", "T02", "T03", "T04"))
scale_shape_manual("Treatment with Study Drug", values = c(16,1), breaks = c("Injected", "Not Injected"))
scale_alpha_manual("", values = c(1,1), breaks = c("PD-1", "PD"), labels = c("Anti-PD-1 Treatment", "Confirmed PD"))
scale_linetype_manual("", values = c("dashed"), labels = "Crossover")
guides(color = guide_legend(order = 1),
shape = guide_legend(order = 2),
alpha = guide_legend(order = 3, override.aes = list(
shape = c(17, 7),
color = c(palette[2], "black")
)),
linetype = guide_legend(order = 4))
coord_cartesian(xlim = c(-1,6), ylim = c(-12,70))
theme(plot.caption = element_text(size = 8))