Hi all I'm attempting to simulate data that looks something like this: 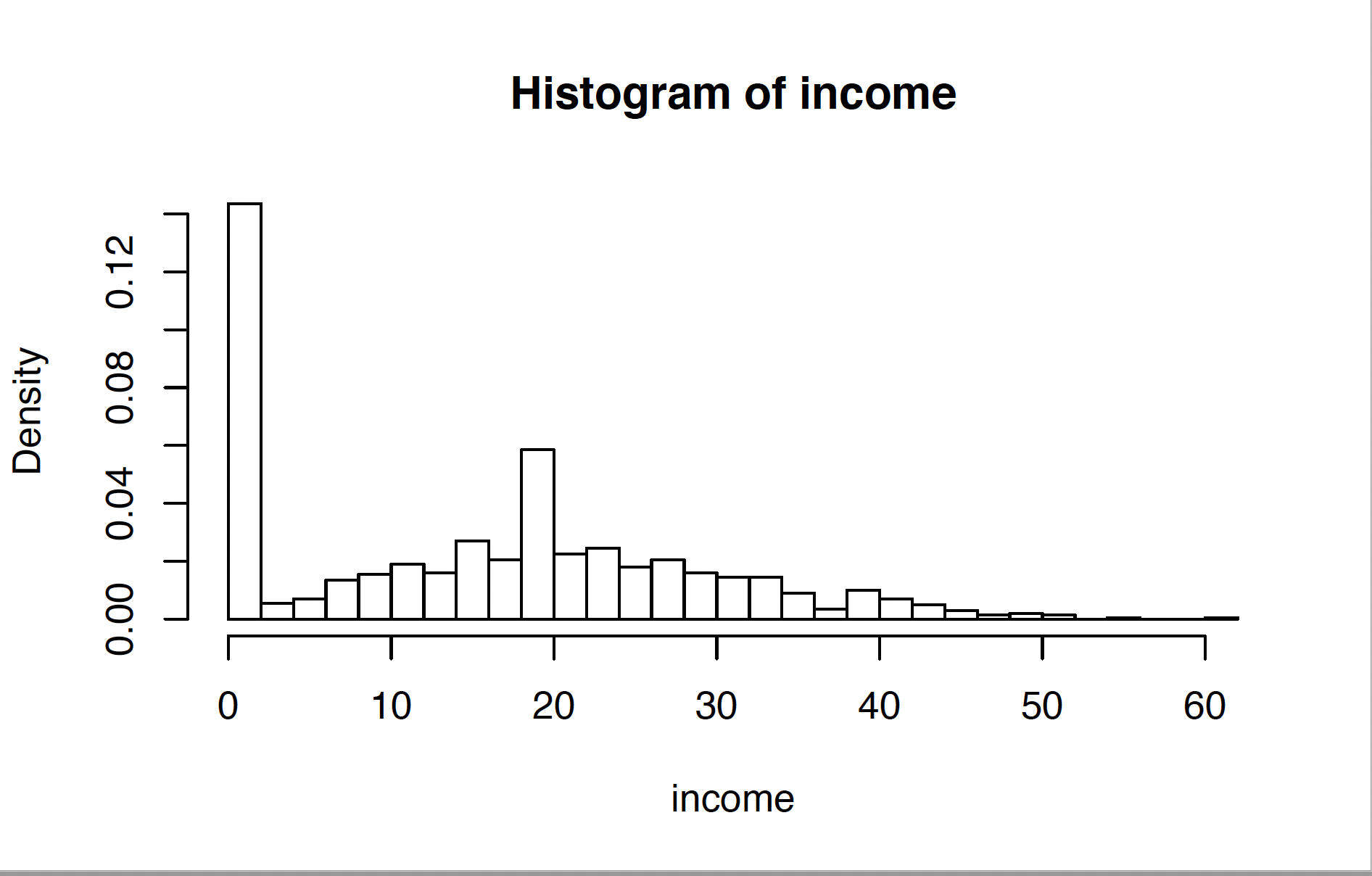
Anyone know how I'd go about doing this?
CodePudding user response:
This can be considered as a classical problem to transform uniform random numbers generated with runif with an inverse transformation from empirical relative frequencies. Here an approach that uses approx:
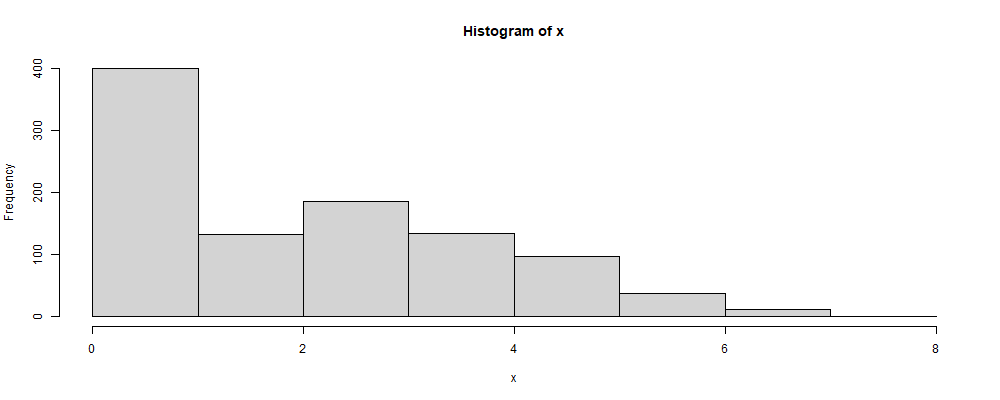
freq <- c(0.39, 0.02, 0.15, 0.18, 0.12, 0.09,0.04, 0.01)
sum(freq) # This is a check. The sum must be 1.0.
r_empirical <- function(n, freq) {
approx(c(0, cumsum(freq)), 0:(length(freq)),
runif(n), method="constant", f=0)$y
}
x <- r_empirical(1000, freq)
hist(x, breaks=0:length(freq))
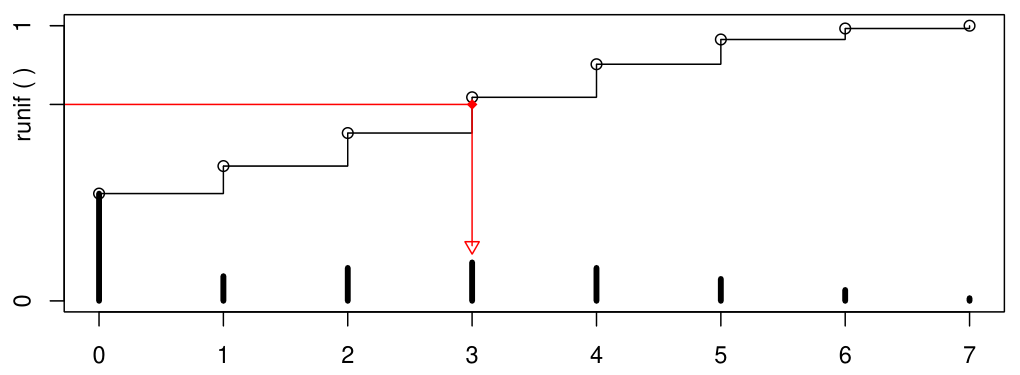
The following figure demonstrates the basic principle. The stairs show the cumulative distribution, the red arrows how a uniform random number can be transformed:
CodePudding user response:
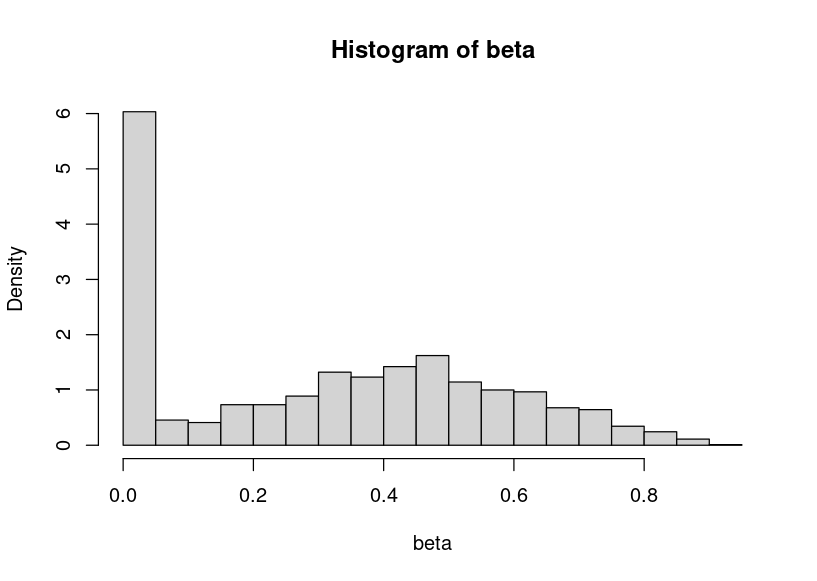
you could also use two beta distributions.
Beta is a very useful distribution.
beta<-c(rbeta(600, 0.1, 5, ncp = 0),rbeta(1200, 3, 4, ncp = 1))
hist(beta,breaks=30,probability = T)
CodePudding user response:
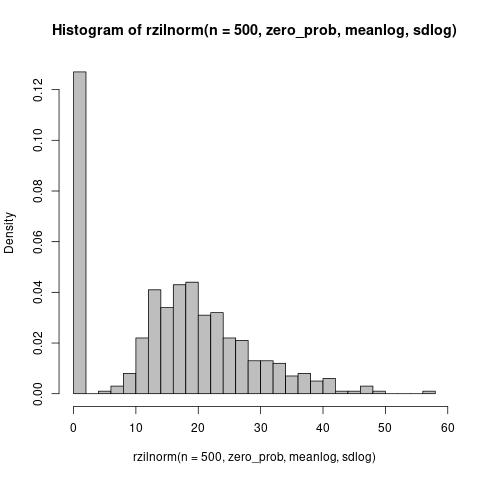
Zero-inflated log-Normal? (Spike at zero looks a little too big for a Tobit, i.e. censored Normal with the negative stuff piled up on zero)
zero_prob <- 0.25
meanlog <- log(20)
sdlog <- 0.4 ## SD ~ 40%
n <- 500
rzilnorm <- function(n, pz, meanlog, sdlog) {
ifelse(runif(n) < pz, 0,
rlnorm(n, meanlog, sdlog))
}
set.seed(101)
hist(rzilnorm(n=500, zero_prob, meanlog, sdlog), col = "gray", breaks=25, freq=FALSE)
My first try was with n=100 and pz=0.2; if I were going to play around with this more I might increase sdlog a little bit. Otherwise this looks pretty close?