Simulate some data
n <- 50
beta0 <- 2
beta1 <- 0.32
x <- runif(n=n, min=0, max=5)
mu <- exp(beta0 beta1 * x)
y <- rpois(n=n, lambda=mu)
data <- data.frame(x=x, y=y)
plot(data$x, data$y)
Apply a Poisson regression
glm.pois <- glm(formula= y ~ x, data=data, family=poisson(link="log"))
summary(glm.pois)
Compute profile 95% CI
(pCI <- confint(glm.pois))
nice.xs <- sort(data$x)
pred.pCI <- apply(t(pCI), 1, FUN=function(x){exp( x[1] x[2]*nice.xs)})
Compute 95% CI using the SE
new.dat <- data.frame(phat = predict(glm.pois, type="response"),
x = data$x)
new.dat <- new.dat[with(new.dat, order(x)), ]
ilink <- poisson()$linkinv
## Add fit and se.fit on the link scale
new.dat <- bind_cols(new.dat,
setNames(as_tibble(predict(glm.pois, new.dat,
se.fit = TRUE)[1:2]),
c('fit_link','se_link')))
new.dat <- mutate(new.dat,
fit = ilink(fit_link),
right_upr = ilink(fit_link (2 * se_link)),
right_lwr = ilink(fit_link - (2 * se_link)))
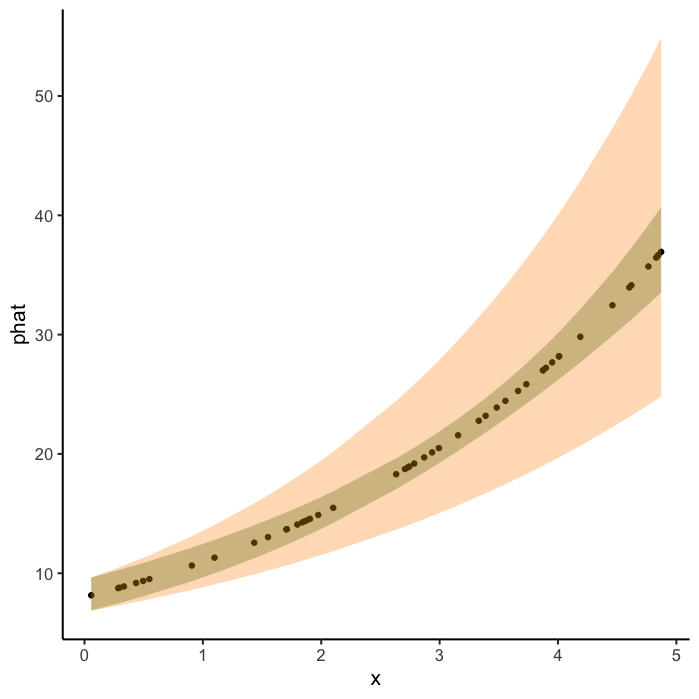
Plot
ggplot(data=new.dat, aes(x=x, y= phat))
geom_point()
geom_ribbon(aes(x=x, ymin =right_lwr, ymax = right_upr),
fill = "darkgreen", alpha=0.3)
geom_ribbon(aes(x=nice.xs, ymin = pred.pCI[,1], ymax = pred.pCI[,2]),
fill = "darkorange", alpha=0.3)
theme_classic(base_size=15)
My question is why are the profile 95% CI (orange ribbon) so large? I imagine I am computing them wrong but I don't know where my mistake is.
CodePudding user response:
There is a very strong correlation between the intercept and slope:
cov2cor(vcov(glm.pois))
(Intercept) x
(Intercept) 1.0000000 -0.9147526
x -0.9147526 1.0000000
Your profile CIs, which are computing (logistic(intercept_lwr slope_lwr*x) and logistic(intercept_upr slope_upr*x), are ignoring this correlation.
It's not easy to compute profile confidence intervals on derived values (i.e., on quantities other than the parameters themselves).