I have a very simple (albeit large) data frame with 2 numeric columns and 1 character grouping column, containing several NAs.
I am going to use iris as an example. Below, I just introduce random NAs in the Species column I want to use for grouping and coloring.
What I do here is to remake the Species column as a factor with "NA" (character) at the end. I make a palette with gray at the end, that I want to correspond to "NA".
data("iris")
set.seed(123)
na_rows <- sample(nrow(iris), 100, replace = F)
iris$Species <- as.character(iris$Species)
iris$Species[na_rows] <- "NA"
mylevels <- iris$Species[which(iris$Species!="NA")]
mylevels <- c(gtools::mixedsort(unique(mylevels)), "NA")
iris$Species <- factor(iris$Species, levels=mylevels)
plot_palette <- c("red","blue","green")
plot_palette <- c(plot_palette[1:length(mylevels)-1], "gray")
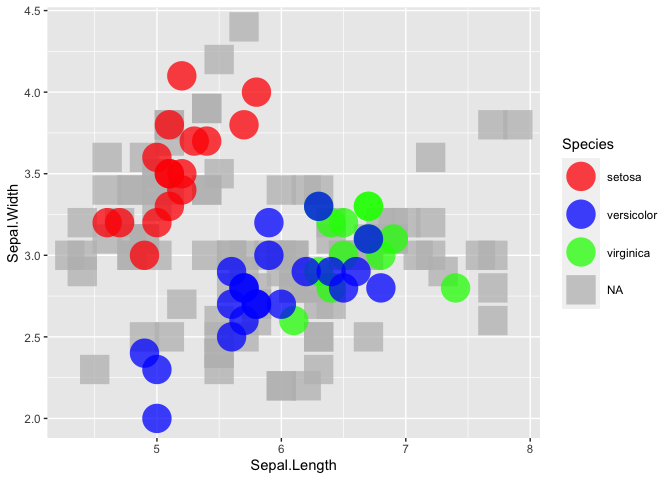
All good till here. Now I make my scatter plot like this:
grDevices::pdf(file="test1.pdf", height=10, width=10)
P <- ggplot2::ggplot(data=iris, ggplot2::aes(x=Sepal.Length, y=Sepal.Width, color=Species))
ggplot2::scale_color_manual(values=plot_palette)
P1 <- P ggplot2::geom_point(pch=16, size=10, alpha=0.75)
print(P1)
grDevices::dev.off()
This produces this plot:
Still all good till here. This is very close to what I want, but my actual data frame is very large, and many non-NA points are hidden behind the NA ones.
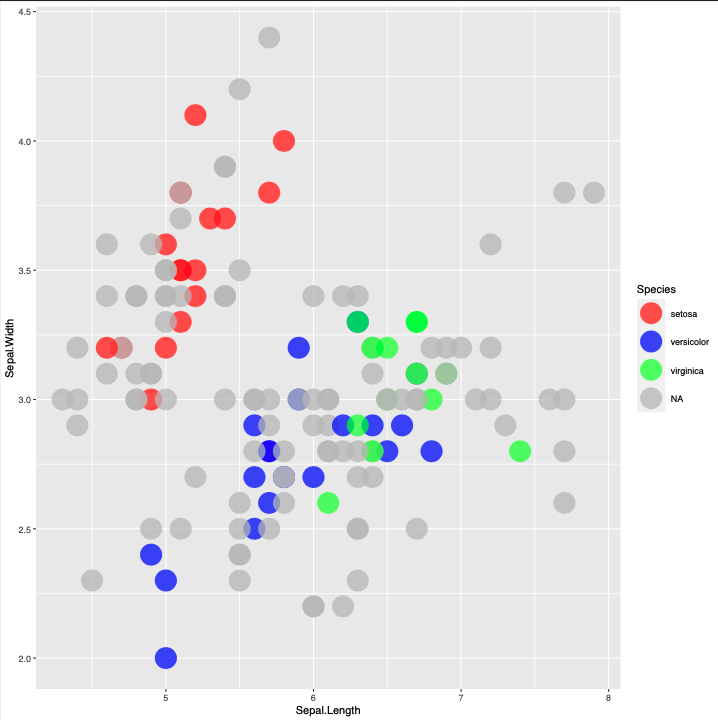
To avoid this, I am trying to plot first the subset of NA data, and then on an upper layer the subset of non-NA data. I try the code below:
grDevices::pdf(file="test2.pdf", height=10, width=10)
P <- ggplot2::ggplot(data=iris, ggplot2::aes(x=Sepal.Length, y=Sepal.Width, color=Species))
ggplot2::scale_color_manual(values=plot_palette)
P1 <- P ggplot2::geom_point(data=function(x){x[x$Species == "NA", ]}, pch=15, size=10, alpha=0.75)
ggplot2::geom_point(data=function(x){x[x$Species != "NA", ]}, pch=16, size=10, alpha=0.75)
print(P1)
grDevices::dev.off()
This produces this plot:
The problem I have here is very obvious, but I have no clue how to solve it.
I just want this second plot to be exactly like the first one, except for the "layering" with NA points behind. I want to maintain the original order of the Species levels in the legend, with NA at the end, and the same color correspondence, with NA associated to gray.
Notice I also changed the pch for NA points. A bonus would be to have the legend with just square for NA (at the bottoms), and just circles for the other samples.
Any help? Thanks!
CodePudding user response:
There is no need for multiple layers. You could simply reorder your dataset so that the NAs get plotted first and for the shapes you could map Species on the shape aes and set the desired shape via scale_shape_manual:
iris1 <- dplyr::arrange(iris, desc(Species))
P <- ggplot2::ggplot(data=iris1, ggplot2::aes(x=Sepal.Length, y=Sepal.Width, color=Species, shape = Species))
ggplot2::scale_color_manual(values=plot_palette)
P ggplot2::geom_point(size=10, alpha=0.75) ggplot2::scale_shape_manual(values = c(16, 16, 16, 15))