I have this code below:
set.seed(1)
n <- 100
x <- rnorm(n)
y <- 5 3*x rnorm(n, mean = 0, sd = 0.3)
df <- data.frame(x=x,y=y)
slope <- lm(y~x, data = df)$coefficients[2]
I want to use 100 bootstrap samples to estimate a 95% confidence interval for the slope coefficient. I've never done bootstrapping before so I'm a little stuck. I know I want to use sample to sample 100 indices from 1:n with replacement. I also know I need the standard error of the slope to calculate CI. However, when I do stderr<- sd(slope)/sqrt(length(slope)) I get <NA>. Just not sure how to translate that into R. I should get the CIs to be (2.95, 3.05).
CodePudding user response:
A single bootstrap sample of c(1, 2, 3, 4, 5) can be sampled via sample(c(1, 2, 3, 4, 5), 5, TRUE). Many can be drawn using replicate on that:
# this was your original code
set.seed(1)
n <- 100
x <- rnorm(n)
y <- 5 3*x rnorm(n, mean = 0, sd = 0.3)
df <- data.frame(x=x,y=y)
# before we do many bootstrap samples, first define what happens in each one of them
one_regr <- function(){
mysample <- sample(1:n, n, TRUE) # which observations pairs are in this
# bootstrap sample?
my_df <- df[mysample, ]
slope <- lm(y ~x, data = my_df)$coefficients[2]
return(slope)
}
# test run -- works!
one_regr()
# now we can do many replicates with replicate()
many_slopes <- replicate(5000, one_regr())
hist(many_slopes, breaks = 30)
quantile(many_slopes, c(.025, .975))
Addition 1:
You can have more fun with this if you sample not only the slopes but the intercepts as well as in
one_regr <- function(){
mysample <- sample(1:n, n, TRUE) # which observation pairs are in this
# bootstrap sample?
my_df <- df[mysample, ]
return( lm(y ~x, data = my_df)$coefficients )
}
many_lines <- replicate(5000, one_regr())
intercepts <- many_lines[1,]
slopes <- many_lines[2,]
plot(df$x, df$y, main = "data and 500 regression lines")
for(i in 1:5000){ # just draw the regression lines
abline(a = intercepts[i], b = slopes[i], col = "#00009001")
}
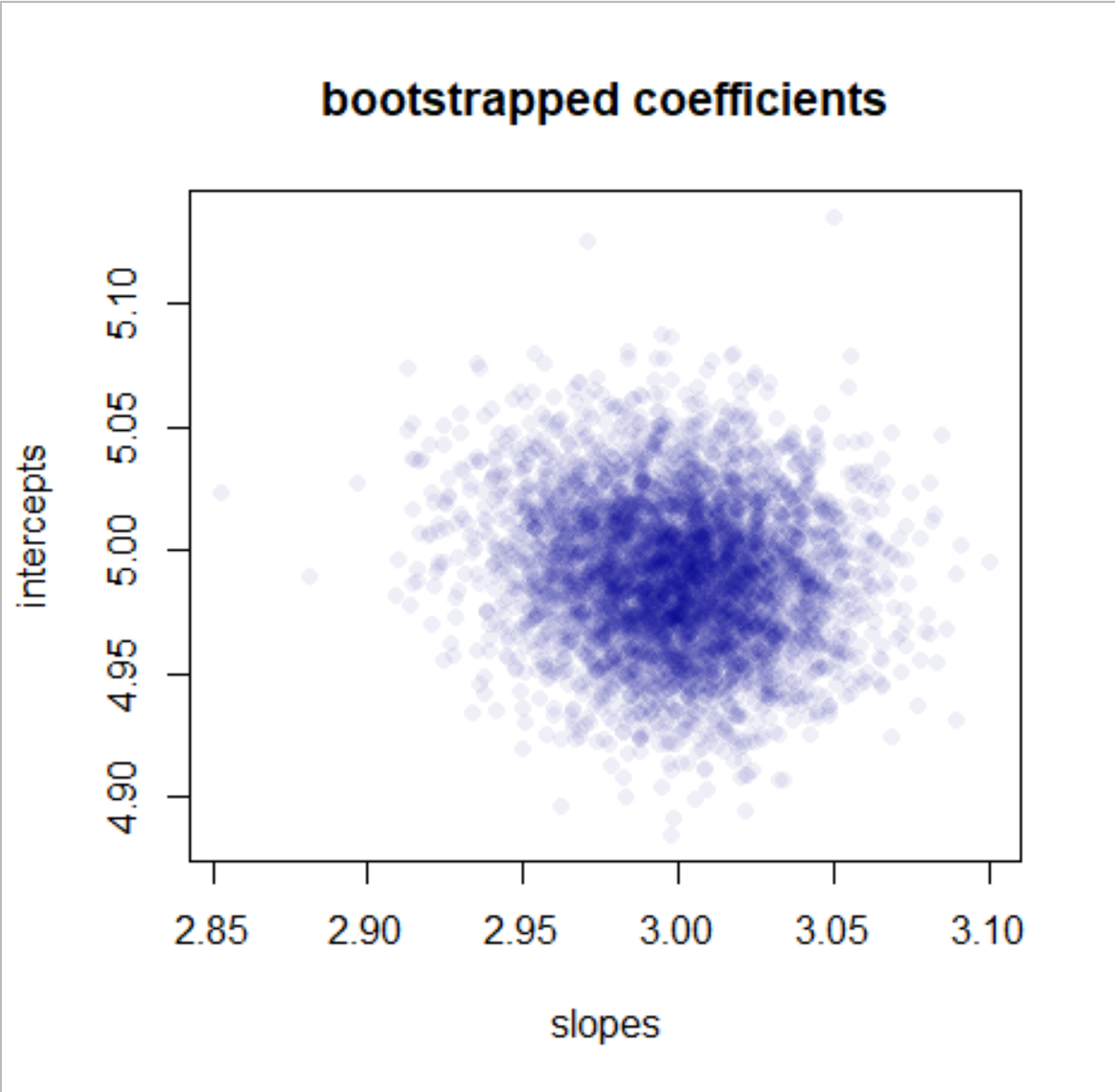
plot(slopes, intercepts, main = "bootstrapped coefficients",
pch = 16, col = "#00009010")
quantile(slopes, c(.025, .975))
Addition 2:
https://www.investopedia.com/terms/s/standard-error.asp states:
The standard error (SE) of a statistic is the approximate standard deviation of a statistical sample population.
So the standard error of the slope is the standard deviation of our bootstrapped slopes:
> sd(slopes)
[1] 0.02781162
roughly, this should be 4 times the width of the 95%-CI:
> sd(slopes)
[1] 0.02781162
> abs(2.95 - 3.05)/4
[1] 0.025