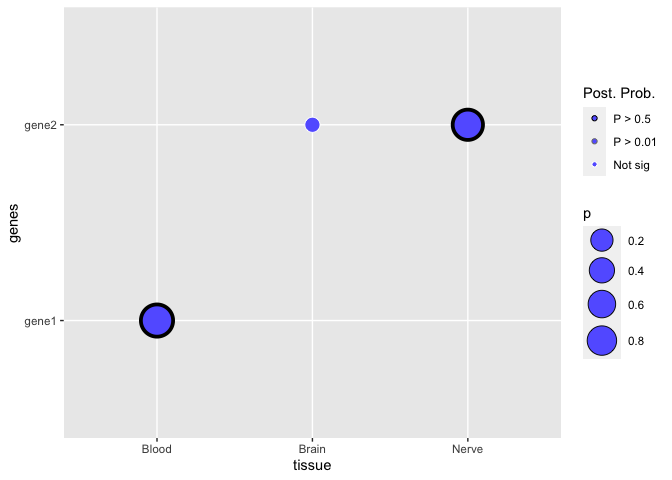
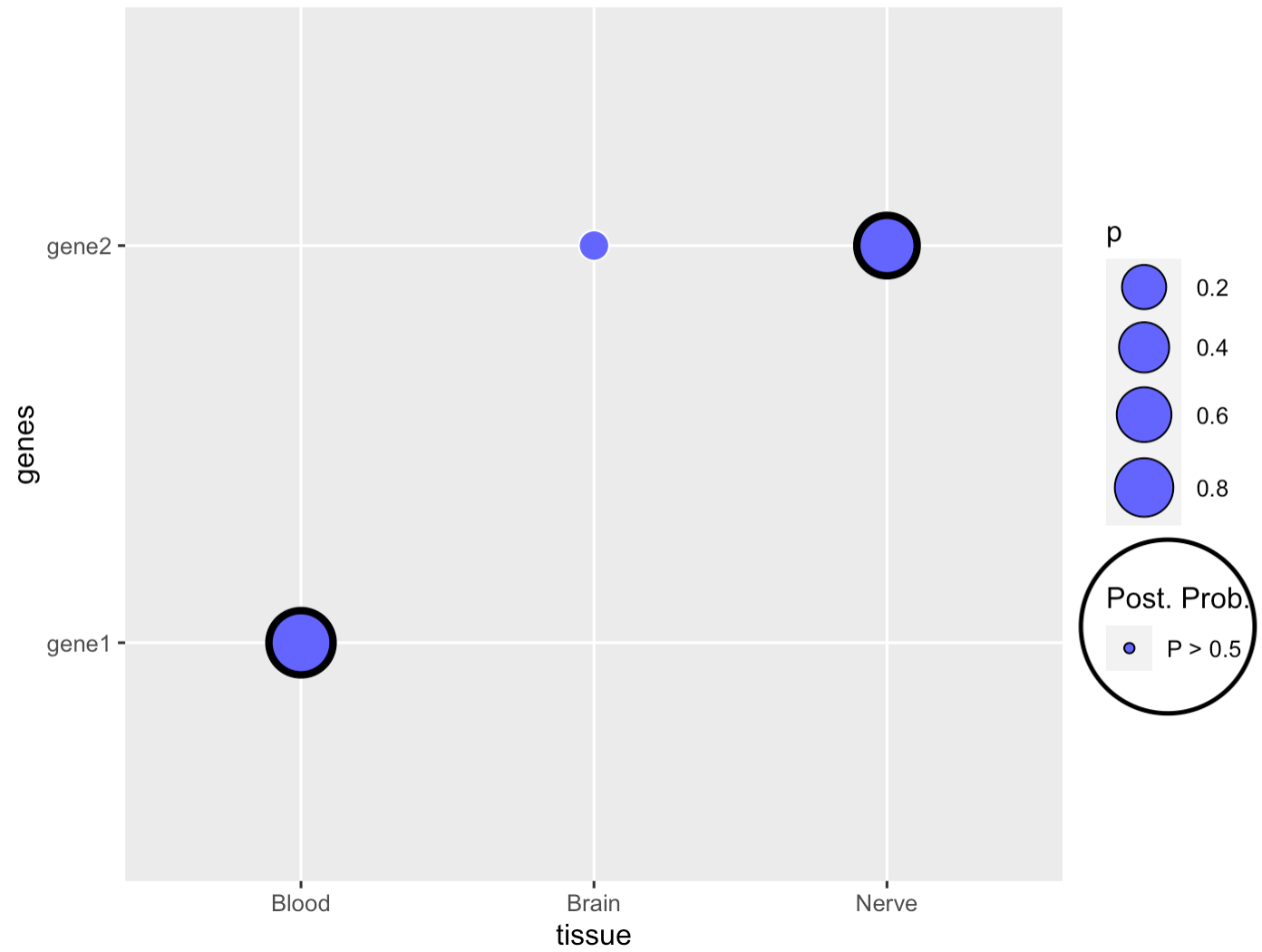
Suppose I have this dataframe:
# packages
library(ggplot2)
library(dplyr)
# data
genes <- c("gene1", "gene2", "gene2")
tissue <- c("Blood", "Nerve", "Brain")
pval <- c(0.8, 0.6, 0.005)
df <- data.frame(col_x = tissue,
col_y = genes,
p = pval)
I create a column for labeling purposes in the plot:
df <- df %>%
mutate(`Post. Prob.` = ifelse(`p`>= 0.5, "P > 0.5",
ifelse(`p`>= 0.01 & `p`<= 0.5, "P > 0.01","Not sig")))
The remaining code is for different aesthetics in the plot.
# black ring if P>0.5
# gray ring if between 0.01 and 0.5
# white ring if not sig
ring_df <- df %>%
select(`Post. Prob.`) %>%
mutate(`ring` = ifelse(`Post. Prob.` == "P > 0.5", "black",
ifelse(`Post. Prob.` == "P > 0.01", "gray", "white")))
rings <- ring_df$ring
names(rings) <- ring_df$`Post. Prob.`
Now I create the plot:
ggplot(df)
geom_point(aes(x=`col_x`, y=`col_y`,
size=`p`,
color=`Post. Prob.`),
shape=21, # so I can fill and change the border color
fill="#6565ff", # light blue
stroke = ifelse(df$`Post. Prob.` == "P > 0.5",2, # stroke is the thickness of the border
ifelse(df$`Post. Prob.` == "P > 0.01", 2, 0.5)))
scale_color_manual(values=rings,
breaks=c("P > 0.5", "P > 0.01", "Not Sig."),
labels=c("P > 0.5", "P > 0.01", "Not Sig."),
name="Post. Prob.")
scale_size(range=c(5,10))
xlab("tissue")
ylab("genes")
However, my question is that in the legend, the plot only shows P > 0.5. I know that none of the points follow the other parameters, but how do I make it so that the gray legend marker would show?
CodePudding user response:
One option to achieve your desired result would be
- to convert your post prob column to a factor with the appropriate levels
- set
drop=FALSEandlimits=forceinsidescale_color_manualto avoid that unused factor levels get dropped
Notes:
- Instead of using nested
ifelseI would suggest to switch tocase_when. - There is no need to wrap each column name inside backticks. Backticks are only needed if you use awkward column names including e.g. spaces.
- I would suggest to avoid using column names including spaces such as
Post. Prob.as there are easier ways to get nice labels e.g. usinglabs()
library(ggplot2)
library(dplyr)
genes <- c("gene1", "gene2", "gene2")
tissue <- c("Blood", "Nerve", "Brain")
pval <- c(0.8, 0.6, 0.005)
df <- data.frame(
col_x = tissue,
col_y = genes,
p = pval
)
df <- df %>%
mutate(post_prob = case_when(
p >= 0.5 ~ "P > 0.5",
p >= 0.01 & p <= 0.5 ~ "P > 0.01",
TRUE ~ "Not sig"),
post_prob = factor(post_prob, levels = c("P > 0.5", "P > 0.01", "Not sig"))
)
rings <- df %>%
select(post_prob) %>%
mutate(ring = case_when(
post_prob == "P > 0.5" ~ "black",
post_prob == "P > 0.01" ~ "gray",
TRUE ~ "white")
) %>%
tibble::deframe()
ggplot(df)
geom_point(aes(
x = col_x, y = col_y,
size = p,
color = post_prob,
stroke = case_when(
post_prob == "P > 0.5" ~ 2,
post_prob == "P > 0.01" ~ 2,
TRUE ~ 0.5
)
),
shape = 21,
fill = "#6565ff"
)
scale_color_manual(
values = rings,
name = "Post. Prob.",
drop = FALSE,
limits = force
)
scale_size(range = c(5, 10))
xlab("tissue")
ylab("genes")