I have the following data frame regarding a predator, prey interaction, SS=surf.smelt, SL = sandlance and H = herring.
Essentially all I need is another column that states whether there was more than one species available for the predator during the interaction. For example, If you look at index 12, the Prey was SL, but there was 1000 sandlance and 100 herring available, I need a column that can simply show with a 1 or 0 if there were more than 2 species available for the predator.
If possible, I would also like to show what other species was available in long format
Date frame I have:
index date site Pred Prey attack passive surf.smelt sandlance herring
17 2015-06-06 cb JCK SS 0 1 20 0 0
26 2015-07-05 cb JCK SS 0 1 100 0 0
12 2016-07-26 cb JCK SL 1 0 0 1000 100
88 2016-07-26 cb JCK H 1 0 0 1000 1000
89 2016-07-26 cb JCK H 1 0 0 0 100
90 2018-08-21 cb JCO SL 1 0 100 500 0
100 2018-08-26 cb JCO SL 1 0 0 1000 100
108 2019-06-22 cb JCO SS 0 1 50 0 100
Data frame I want:
index date site Pred Prey attack passive surf.smelt sandlance herring OtherPrey?
17 2015-06-06 cb JCK SS 0 1 20 0 0 0
26 2015-07-05 cb JCK SS 0 1 100 0 0 0
12 2016-07-26 cb JCK SL 1 0 0 1000 100 1
88 2016-07-26 cb JCK H 1 0 0 1000 1000 1
89 2016-07-26 cb JCK H 1 0 0 0 100 0
90 2018-08-21 cb JCO SL 1 0 100 500 0 1
100 2018-08-26 cb JCO SL 1 0 0 1000 100 1
108 2019-06-22 cb JCO SS 0 1 50 0 100 1
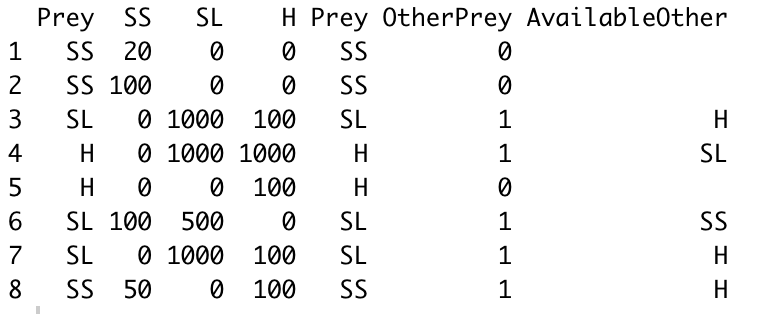
And if possible I would want to define the other species available:
Data frame I want:
index date site Pred Prey attack passive surf.smelt sandlance herring OtherAvailable
17 2015-06-06 cb JCK SS 0 1 20 0 0 0
26 2015-07-05 cb JCK SS 0 1 100 0 0 0
12 2016-07-26 cb JCK SL 1 0 0 1000 100 H
88 2016-07-26 cb JCK H 1 0 0 1000 1000 SL
89 2016-07-26 cb JCK H 1 0 0 0 100 0
90 2018-08-21 cb JCO SL 1 0 100 500 0 SS
100 2018-08-26 cb JCO SL 1 0 0 1000 100 H
108 2019-06-22 cb JCO SS 0 1 50 0 100 H
CodePudding user response:
With dplyr:
df %>%
rowwise() %>%
mutate(Other_Prey=ifelse(sum(c_across(surf.smelt:herring)>0)>=2,1,0))
# A tibble: 8 x 11
# Rowwise:
index date site Pred Prey attack passive surf.smelt sandlance herring Other_Prey
<int> <chr> <chr> <chr> <chr> <int> <int> <int> <int> <int> <dbl>
1 17 2015-06-06 cb JCK SS 0 1 20 0 0 0
2 26 2015-07-05 cb JCK SS 0 1 100 0 0 0
3 12 2016-07-26 cb JCK SL 1 0 0 1000 100 1
4 88 2016-07-26 cb JCK H 1 0 0 1000 1000 1
5 89 2016-07-26 cb JCK H 1 0 0 0 100 0
6 90 2018-08-21 cb JCO SL 1 0 100 500 0 1
7 100 2018-08-26 cb JCO SL 1 0 0 1000 100 1
8 108 2019-06-22 cb JCO SS 0 1 50 0 100 1
CodePudding user response:
We can use dplyr::if_any:
library(dplyr)
df %>% mutate(other_prey = if_any(surf.smelt:herring))
For the "OtherAvailable", we can use toString
library(dplyr)
df %>% rowwise %>%
mutate(OtherAvailable = toString(names(across(surf.smelt:herring))[as.logical(surf.smelt:herring)]))
CodePudding user response:
It's not very elegant, but here's a shot.
library(dplyr)
library(stringr)
df <- data.frame(Prey = c("SS", "SS", "SL", "H", "H", "SL", "SL", "SS"),
SS = c(20, 100, 0, 0, 0, 100, 0 ,50),
SL = c(0,0,1000, 1000, 0, 500, 1000, 0),
H = c(0,0,100,1000,100,0,100,100) )
tb <- df %>%
mutate(SS = SS > 0, SL = SL > 0, H = H > 0,
OtherPrey = (rowSums(across(where(is.logical))))-1)
tb1 <- which(as.matrix(tb[,2:4]), arr.ind = TRUE)
tb$Available <- tapply(names(tb[,2:4])[tb1[,2]], tb1[,1], paste, collapse=",")
tb <- tb %>%
mutate(other = str_replace(Available,Prey,""),
AvailableOther = str_replace(other,",","")) %>%
select(Prey, OtherPrey, AvailableOther)
df_new <- cbind(df, tb)
df_new