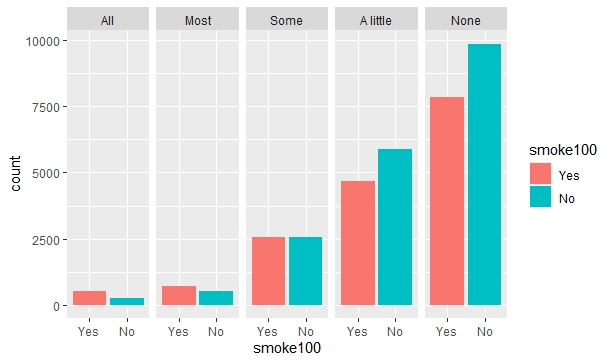
There seems to be a problem with R's ggplot2 library when I include both the fill and group parameters in a bar plot (geom_bar()). I've already tried looking for answers for several hours but couldn't find one that would help. This is actually my first post here.
To give a little background, I have a dataframe named smokement (short for smoke and mental health), a categorical variable named smoke100 (smoked in the past 100 days?) with "Yes" and "No", and another categorical variable named misnervs (frequency of feelings of nervousness) with 5 possible values: "All", "Most", "Some", "A little", and "None."
When I run this code, I get this result:
ggplot(data = smokement)
geom_bar(aes(x = smoke100, fill = smoke100))
facet_wrap(~misnervs, nrow = 1)
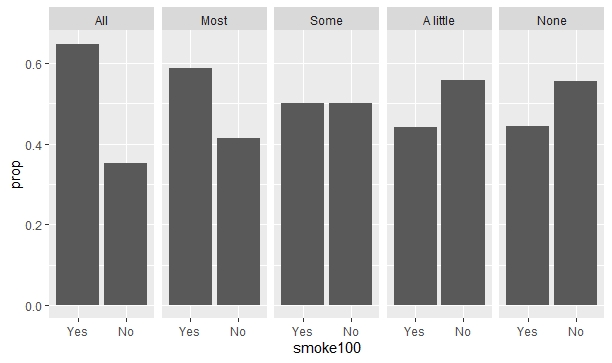
However, the result I want is to have all grouped bar plots display their respective proportions. By reading a bit of "R for Data Science" book I found out that I need to include y = ..prop.. and group = 1 in aes() to achieve it:
ggplot(data = smokement)
geom_bar(aes(x = smoke100, y = ..prop.., group = 1))
facet_wrap(~misnervs, nrow = 1)
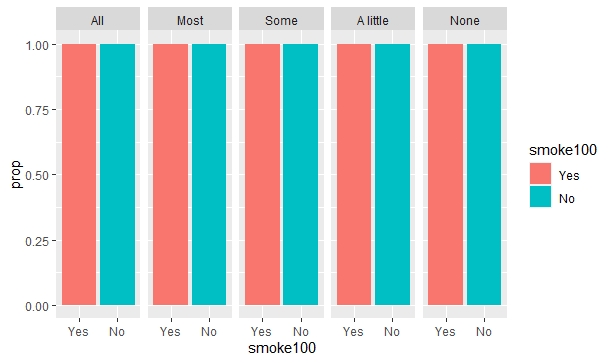
Finally, I try to use the fill = smoke100 parameter in aes() to display this categorical variable in color, just like I did on the first code. But when I add this fill parameter, it doesn't work! The code runs, but it shows exactly the same output as the second code, as if the fill parameter this time was somehow ignored!
ggplot(data = smokement)
geom_bar(aes(x = smoke100, y = ..prop.., group = 1, fill = smoke100))
facet_wrap(~misnervs, nrow = 1)
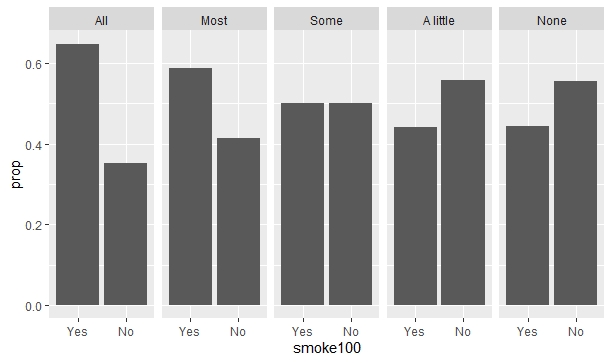
Does anyone have an idea of why this happens, and how to solve it? My end goal is to display each value of smoke100 (the "Yes" and "No" bars) with colors and a legend at the right, just like on the first graph, while having each grouping level of "misnervs" display their respective proportions of smoke100 ("Yes", "No") levels, just like on the second graph.
EDIT:
> dim(smokement)
[1] 35471 6
> str(smokement)
'data.frame': 35471 obs. of 6 variables:
$ smoke100: Factor w/ 2 levels "Yes","No": 1 2 1 2 1 1 1 1 1 1 ...
$ misnervs: Factor w/ 5 levels "All","Most","Some",..: 3 4 5 4 1 5 3 3 5 5 ...
$ mishopls: Factor w/ 5 levels "All","Most","Some",..: 3 5 5 5 5 5 5 5 5 5 ...
$ misrstls: Factor w/ 5 levels "All","Most","Some",..: 3 5 5 3 1 5 3 5 1 5 ...
$ misdeprd: Factor w/ 5 levels "All","Most","Some",..: 5 5 5 5 4 5 5 5 5 5 ...
$ miswtles: Factor w/ 5 levels "All","Most","Some",..: 5 5 5 5 5 5 5 5 5 5 ...
> head(smokement)
smoke100 misnervs mishopls misrstls misdeprd miswtles
1 Yes Some Some Some None None
2 No A little None None None None
3 Yes None None None None None
4 No A little None Some None None
5 Yes All None All A little None
6 Yes None None None None None
As for the output without group = 1
ggplot(data = smokement)
geom_bar(aes(x = smoke100, y = ..prop.., fill = smoke100))
facet_wrap(~misnervs, nrow = 1)
CodePudding user response:
Besides the solution offered 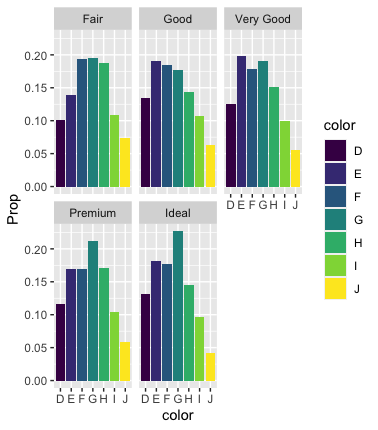 Note the switch from
Note the switch from geom_bar to geom_col: geom_bar uses row counts, geom_col uses values in the data.
As a rough-and-ready QC, here's the equivalent of your code that produces the "all grey' plot:
diamonds %>%
ggplot()
geom_bar(aes(x=color, y=..prop.., fill=color, group=1))
facet_wrap(~cut)