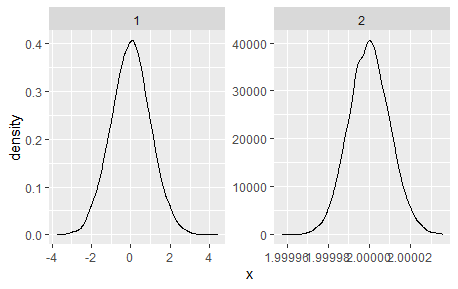
I have a figure created with facet_wrap visualizing the estimated density of many groups. Some of the groups have a much smaller variance than others. This leads to the x axis not being readable for some panels. Minimum reproducable example:
library(tidyverse)
x1 <- rnorm(1e4)
x2 <- rnorm(1e4,mean=2,sd=0.00001)
data.frame(x=c(x1,x2),group=c(rep("1",length(x1)),rep("2",length(x2)))) %>%
ggplot(.) geom_density(aes(x=x)) facet_wrap(~group,scales="free")
The obvious solution to the problem is to increase the figure size, so that everything becomes readable. However, there are too many panels to make this a useful solution. My favourite solution would be to control the number of axis ticks, for example allow for only two ticks on all x-axes. Is there a way to accomplish this?
Edit after suggestions:
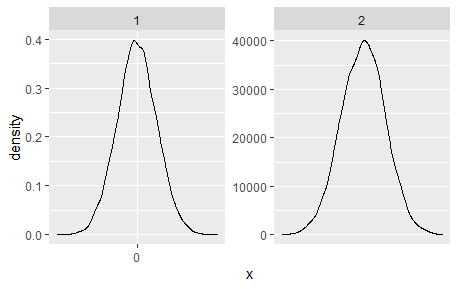
Adding scale_x_continuous(n.breaks = 2) looks like it should exactly do what I want, but it actually does not:
Following the answer in the suggested question 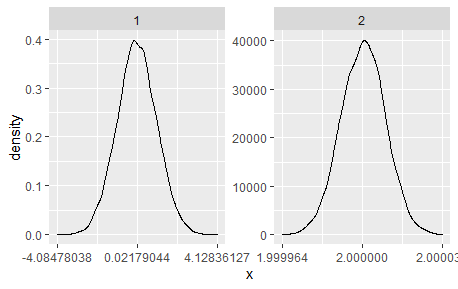
CodePudding user response:
You can add if(seq[2]-seq[1] < 10^(-r)) seq else round(seq, r) to the function equal_breaks developed 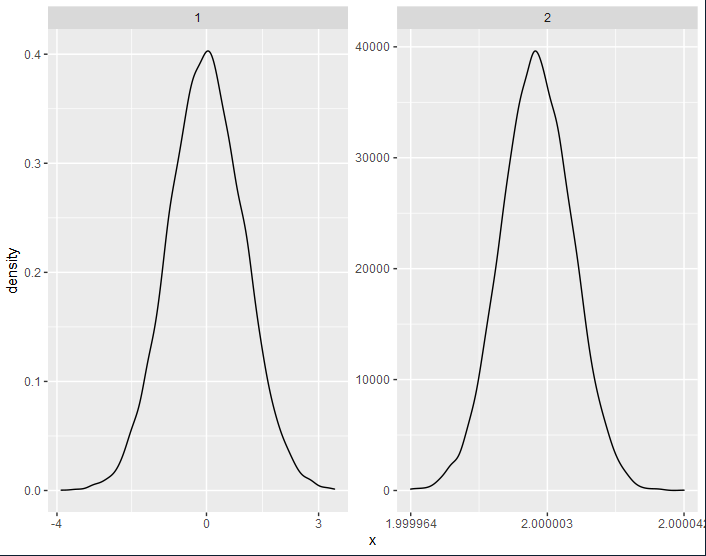
CodePudding user response:
One option to achieve your desired result would be to use a custom breaks and limits function which builds on scales::breaks_extended to get pretty breaks:
library(ggplot2)
set.seed(123)
x1 <- rnorm(1e4)
x2 <- rnorm(1e4,mean=2,sd=0.00001)
mylimits <- function(x) range(scales::breaks_extended()(x))
mybreaks <- function(x) {
breaks <- mylimits(x)
c(breaks, mean(breaks))
}
d <- data.frame(x=c(x1,x2),group=c(rep("1",length(x1)),rep("2",length(x2))))
ggplot(d)
geom_density(aes(x=x))
scale_x_continuous(breaks = mybreaks, limits = mylimits)
facet_wrap(~group,scales="free")
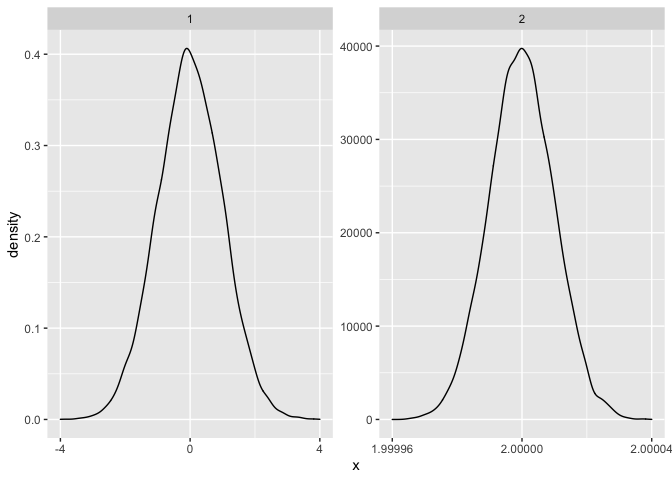
CodePudding user response:
Thanks so much for so many helpful suggestions and great answers! I figured out a solution that works for arbitrarily complex datasets (at least I hope so) by modifying the approach by @Maël and borrowing the great function by RHertel from 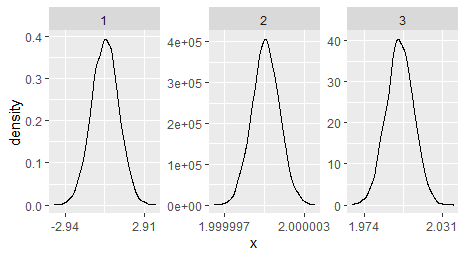
Apologies for answering my own question ... but that would not have been possible without the great help from the community :-)