I have the following table:
Sample UMAP1 UMAP2 colors tissue
C_A1 1.41771557752425 1.22200433730937 yellow2 Urine
C_A2 1.76055361418532 1.34108703383903 yellow2 Urine
C_A3 0.00750224109526362 2.73864657429924 yellow2 Urine
C_A4 1.6500702891979 1.08443364179582 yellow2 Urine
C_A5 1.47433412636331 1.24338667538958 yellow2 Urine
C_A9 1.81676319795816 1.41560757063772 yellow2 Urine
C_B1 1.58085792476313 1.06578750509132 yellow2 Urine
C_B2 0.54619869458667 1.47703917280933 yellow2 Urine
CKD_C1 1.77876535894167 2.1866355905902 yellow2 Urine
CKD_C2 1.5528582393691 1.56720266537748 yellow2 Urine
CKD_C3 0.00527727382088514 2.62619847720534 yellow2 Urine
CKD_C4 0.586308172828281 2.7784840588712 yellow2 Urine
CKD_C5 0.186228467054963 2.6796009437192 yellow2 Urine
PYL_E1 -3.07772498762527 4.45095110253223 yellow2 Urine
PYL_E2 1.1175301960485 1.06349707623584 yellow2 Urine
PYL_E3 1.82042047858787 1.42547383706531 yellow2 Urine
PYL_E4 1.66009595777039 1.72432475907437 yellow2 Urine
PYL_E5 1.13056114256336 1.13760951795915 yellow2 Urine
PYL_E6 1.79463825137017 1.47814506344302 yellow2 Urine
PYL_E7 0.622690635767324 -1.96794491349824 yellow2 Urine
PYL_E8 0.63118627267307 1.29871116070517 yellow2 Urine
PYL_E9 -4.58033059073059 3.96133161881786 yellow2 Urine
PYL_F1 1.51666575038286 1.59237710006909 yellow2 Urine
UO_D1 0.709257524588014 1.24974065107936 yellow2 Urine
UO_D2 1.69833621648934 2.18100899996296 yellow2 Urine
UO_D3 1.01987315310411 1.08349161107457 yellow2 Urine
UO_D4 1.53357549894633 1.46876538726954 yellow2 Urine
UO_D5 0.618372187901223 1.1600432701727 yellow2 Urine
UO_D6 1.80740300575174 1.93028754079873 yellow2 Urine
UO_D7 -1.90935774520858 2.21114064992169 yellow2 Urine
SBU_G1 0.0684949931212032 2.6323213276562 yellow2 Urine
SBU_G2 1.37378744895387 1.15990665575179 yellow2 Urine
SBU_G3 0.681350048991714 1.29504193314325 yellow2 Urine
SBU_G4 0.585511550079965 1.46021542902406 yellow2 Urine
SBU_G5 2.13609698314454 -1.52225184779846 yellow2 Urine
BR1.RPM. -3.28586643197682 3.74476178876053 darkgrey Brain
BR2.RPM. -3.66581644761413 3.78800081614875 darkgrey Brain
BR3.RPM. -3.96475462680686 3.8275429988704 darkgrey Brain
BR4.RPM. -3.6558646156002 3.90023979275899 darkgrey Brain
BR5.RPM. -3.77917418926683 3.65892915155001 darkgrey Brain
CO1.RPM. 1.19249345974165 -1.0914160958934 forestgreen Colon
CO2.RPM. 0.853658643619059 -0.658496441780366 forestgreen Colon
CO3.RPM. 0.975760223303845 -1.28665248361392 forestgreen Colon
CO5.RPM. 1.36511790751794 -2.13567084452923 forestgreen Colon
DU1.RPM. 1.63592949492624 -2.46405219183458 olivedrab4 Duodenum
DU2.RPM. 1.87925711931471 -3.03633498025664 olivedrab4 Duodenum
DU5.RPM. 1.37754729522807 -0.478401566279114 olivedrab4 Duodenum
HE1.RPM. -1.84787050174421 3.29683025222076 hotpink3 Heart
HE2.RPM. -1.57712374669721 3.72779965312866 hotpink3 Heart
HE3.RPM. -1.62595902698841 3.4606008066362 hotpink3 Heart
HE4.RPM. -0.907119493708492 3.17316852377154 hotpink3 Heart
HE5.RPM. -1.65644593797311 3.82195960352629 hotpink3 Heart
IL1.RPM. 1.44153778801558 -1.02823513385223 palegreen3 Ileum
IL2.RPM. 1.4889578583487 -2.2804483432248 palegreen3 Ileum
IL3.RPM. 1.91342848325147 -2.10569098957249 palegreen3 Ileum
IL4.RPM. 0.962529810173936 -0.757115204974117 palegreen3 Ileum
JE1.RPM. 1.93437797225407 -2.65674160788117 darkseagreen Jejunum
JE2.RPM. 2.06996643344844 -2.78640061968243 darkseagreen Jejunum
JE3.RPM. 1.62706829576533 -2.79624294732553 darkseagreen Jejunum
JE4.RPM. 1.0825326758516 -2.76470251848877 darkseagreen Jejunum
JE5.RPM. 1.38342677881686 -2.6755546386966 darkseagreen Jejunum
KI1.RPM. 0.967578089848615 1.67184647007406 orange3 Kidney
KI2.RPM. 1.13378641637209 1.34124080803019 orange3 Kidney
KI3.RPM. 1.37130716354727 1.80996835872441 orange3 Kidney
KI4.RPM. 1.07772033561742 1.64994468114772 orange3 Kidney
KI5.RPM. 1.0955496159798 2.0869739595772 orange3 Kidney
LI1.RPM. 1.682084870402 -6.14052679905332 sienna Liver
LI2.RPM. 1.59906448215381 -6.29131805082633 sienna Liver
LI3.RPM. 1.45802709716468 -6.13448078364028 sienna Liver
LI4.RPM. 1.95665039443957 -6.5130493984287 sienna Liver
LI5.RPM. 1.80607410919614 -6.35273035030961 sienna Liver
LU1.RPM. -2.52491770797571 3.4865019812581 mediumpurple Lung
LU2.RPM. -2.86699310757743 3.34528752430189 mediumpurple Lung
LU3.RPM. -2.89232280631143 2.95189454920787 mediumpurple Lung
LU4.RPM. -2.66332768628549 3.09050672631981 mediumpurple Lung
LU5.RPM. -3.08812503849215 3.25327915431485 mediumpurple Lung
PA1.RPM. 1.29849330321564 -5.33710072865804 dodgerblue Pancreas
PA2.RPM. 1.19873085339097 -5.26535178376946 dodgerblue Pancreas
PA3.RPM. 1.11108191265227 -5.14709232997355 dodgerblue Pancreas
PA5.RPM. 0.895423334114921 -4.88663713272289 dodgerblue Pancreas
PL1.RPM. -1.06236761107225 3.49621079235936 red3 Blood
PL2.RPM. -1.07888887772428 3.04101229356896 red3 Blood
PL3.RPM. -0.844686506146655 3.23004986615368 red3 Blood
PL5.RPM. -0.720030063721917 3.49005127885431 red3 Blood
SKI.RPM. 0.199109542570694 2.44218482056362 pink2 Skin
SM1.RPM. 0.563134748949276 2.02657901642232 plum Muscle
SM2.RPM. 0.340223402531866 2.27754829463276 plum Muscle
SM3.RPM. 0.770024511959072 1.97953000238431 plum Muscle
After loading the table, please remember to sort the colors and tissue factors as required:
Cat_urine$colors <- factor(Cat_urine$colors, levels=c("yellow2","darkgrey", "forestgreen", "olivedrab4", "palegreen3", "darkseagreen", "red3", "hotpink3", "plum", "pink2",
"orange3", "sienna", "mediumpurple", "dodgerblue"))
Cat_urine$tissue <- factor(Cat_urine$tissue, levels=c("Urine","Brain", "Colon", "Duodenum", "Jejunum", "Ileum", "Plasma", "Heart", "Skeletal muscle", "Skin",
"Kidney", "Liver", "Lungs", "Pancreas"))
And the following ggplot2 code:
ggplot(Cat_urine, aes(x=UMAP1, y=UMAP2, color=colors))
geom_point(size=5)
scale_color_manual(labels = c("Urine","Brain", "Colon", "Duodenum", "Jejunum", "Ileum", "Plasma", "Heart", "Skeletal muscle", "Skin",
"Kidney", "Liver", "Lungs", "Pancreas", "Urine"),
values = c("yellow2","darkgrey", "forestgreen", "olivedrab4", "palegreen3", "darkseagreen", "red3", "hotpink3", "plum", "pink2",
"orange3", "sienna", "mediumpurple", "dodgerblue"))
guides(colour = guide_legend(override.aes = list(size=5)))
ylim(c(-7,5)) xlim(c(-5,2.5)) guides(shape = guide_legend(override.aes = list(size = 4)))
theme_classic() theme(axis.text.x = element_text(size=10), axis.title=element_text(size=12),
axis.text.y =element_text(size=10),plot.margin = margin(1, 0.5, 0.5, 1, "cm"))
xlab("\nComponent 1") ylab("Component 2\n")
theme(legend.position = c(0.295, 0.25), legend.title = element_blank(),
legend.background = element_rect(fill = "white", color = "black"))
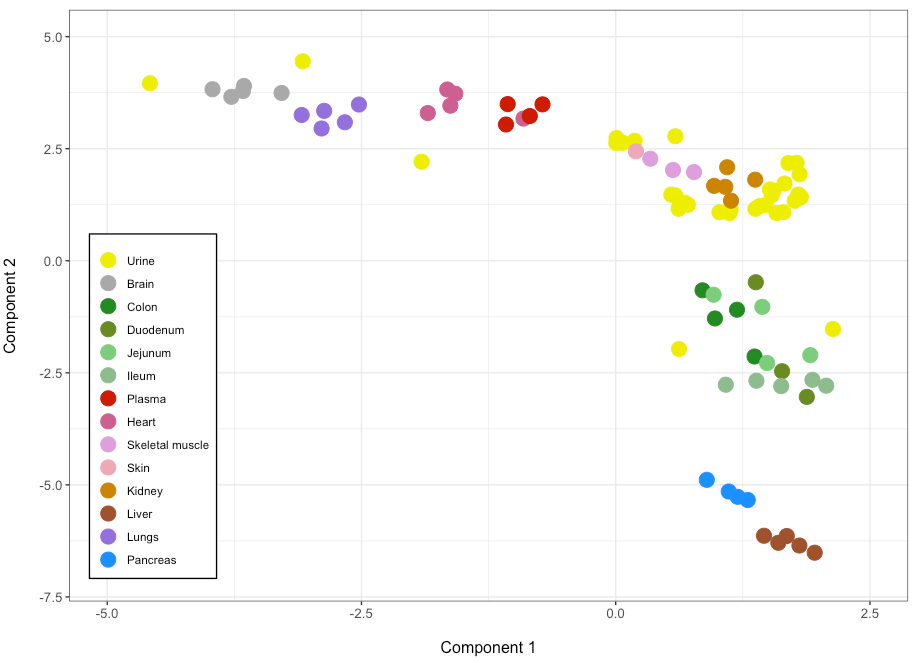
Which generates this figure:
I would like to delimit each dot with a black circular line, and not just the color as it is now, so that the difference between each sample becomes more apparent, mostly focused on those that partially overlap. I tried several ways introducing and changing scale_fill_manual() and scale_color_manual() but it somehow screws up the color assignment...
Could anyone give some hints about it?
Many thanks
CodePudding user response:
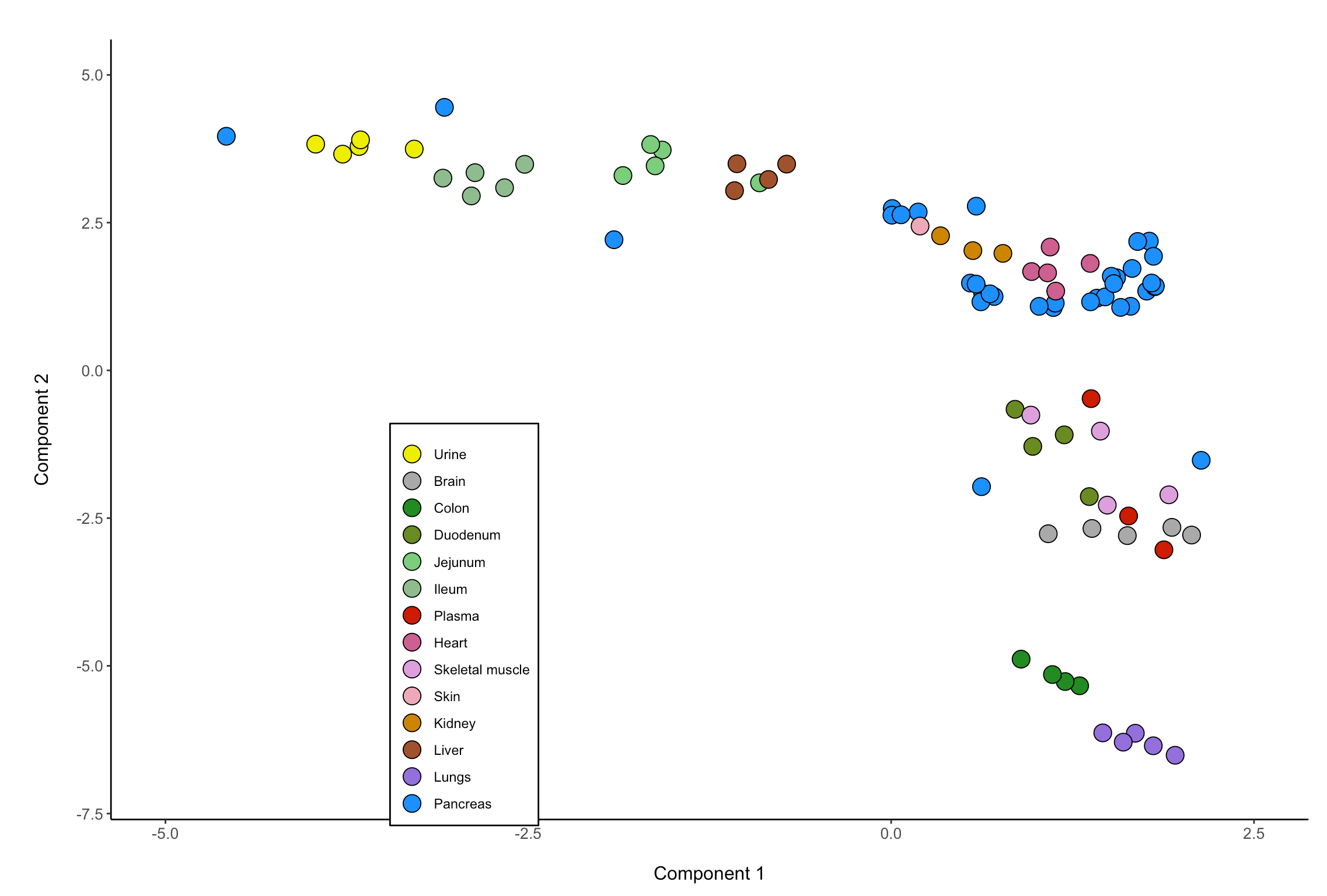
You can add a outline around your dots using the following code. You need to assign a fill to the dots or they will be transparent using fill = colors. I wasn't sure what color outlines you wanted so I gave the dots a black outline. Additionally, if you tried to use the colors from your dataset as the routine colors, ggplot2 will actually realize that the fill and outline are the same color and will try and change the outline. Mainly because having an outline and the fill as the same color would be pointless. This will result in a second legend being made.
One issue here is that you have Urine listed twice as a label so few points are colored yellow2 and the rest are colored with dodgerblue
ggplot(Cat_urine, aes(x=UMAP1, y=UMAP2,fill=colors))
geom_point(size=5, shape=21, color="black")
scale_fill_manual(labels = c("Urine","Brain", "Colon", "Duodenum", "Jejunum", "Ileum", "Plasma", "Heart", "Skeletal muscle", "Skin",
"Kidney", "Liver", "Lungs", "Pancreas", "Urine"),
values = c("yellow2","darkgrey", "forestgreen", "olivedrab4", "palegreen3", "darkseagreen", "red3", "hotpink3", "plum", "pink2",
"orange3", "sienna", "mediumpurple", "dodgerblue"))
guides(colour = guide_legend(override.aes = list(size=5)))
ylim(c(-7,5)) xlim(c(-5,2.5)) guides(shape = guide_legend(override.aes = list(size = 4)))
theme_classic() theme(axis.text.x = element_text(size=10), axis.title=element_text(size=12),
axis.text.y =element_text(size=10),plot.margin = margin(1, 0.5, 0.5, 1, "cm"))
xlab("\nComponent 1") ylab("Component 2\n")
theme(legend.position = c(0.295, 0.25), legend.title = element_blank(),
legend.background = element_rect(fill = "white", color = "black"))
CodePudding user response:
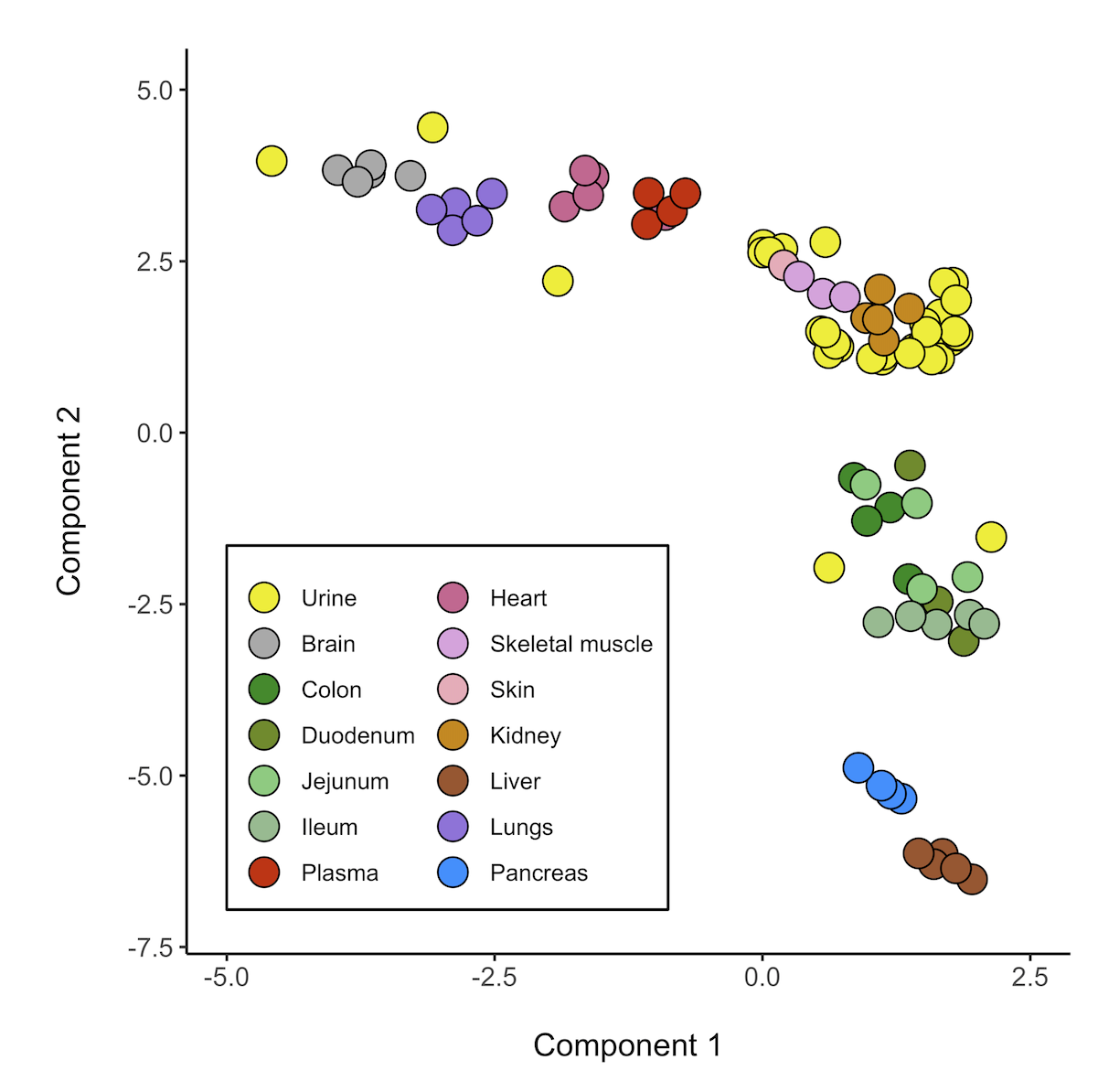
I solved it:
ggplot(Cat_urine, aes(x=UMAP1, y=UMAP2, color="black", fill=colors))
geom_point(size=5, shape=21, color="black")
scale_fill_manual(labels = c("Urine","Brain", "Colon", "Duodenum", "Jejunum", "Ileum", "Plasma", "Heart", "Skeletal muscle", "Skin",
"Kidney", "Liver", "Lungs", "Pancreas"),
values = c("yellow2","darkgrey", "forestgreen", "olivedrab4", "palegreen3", "darkseagreen", "red3", "hotpink3", "plum", "pink2",
"orange3", "sienna", "mediumpurple", "dodgerblue"))
guides(fill = guide_legend(ncol=2, override.aes = list(size=5)))
ylim(c(-7,5)) xlim(c(-5,2.5))
theme_classic() theme(axis.text.x = element_text(size=10), axis.title=element_text(size=12),
axis.text.y =element_text(size=10),plot.margin = margin(1, 0.5, 0.5, 1, "cm"))
xlab("\nComponent 1") ylab("Component 2\n")
theme(legend.position = c(0.295, 0.25), legend.title = element_blank(),
legend.background = element_rect(fill = "white", color = "black"))