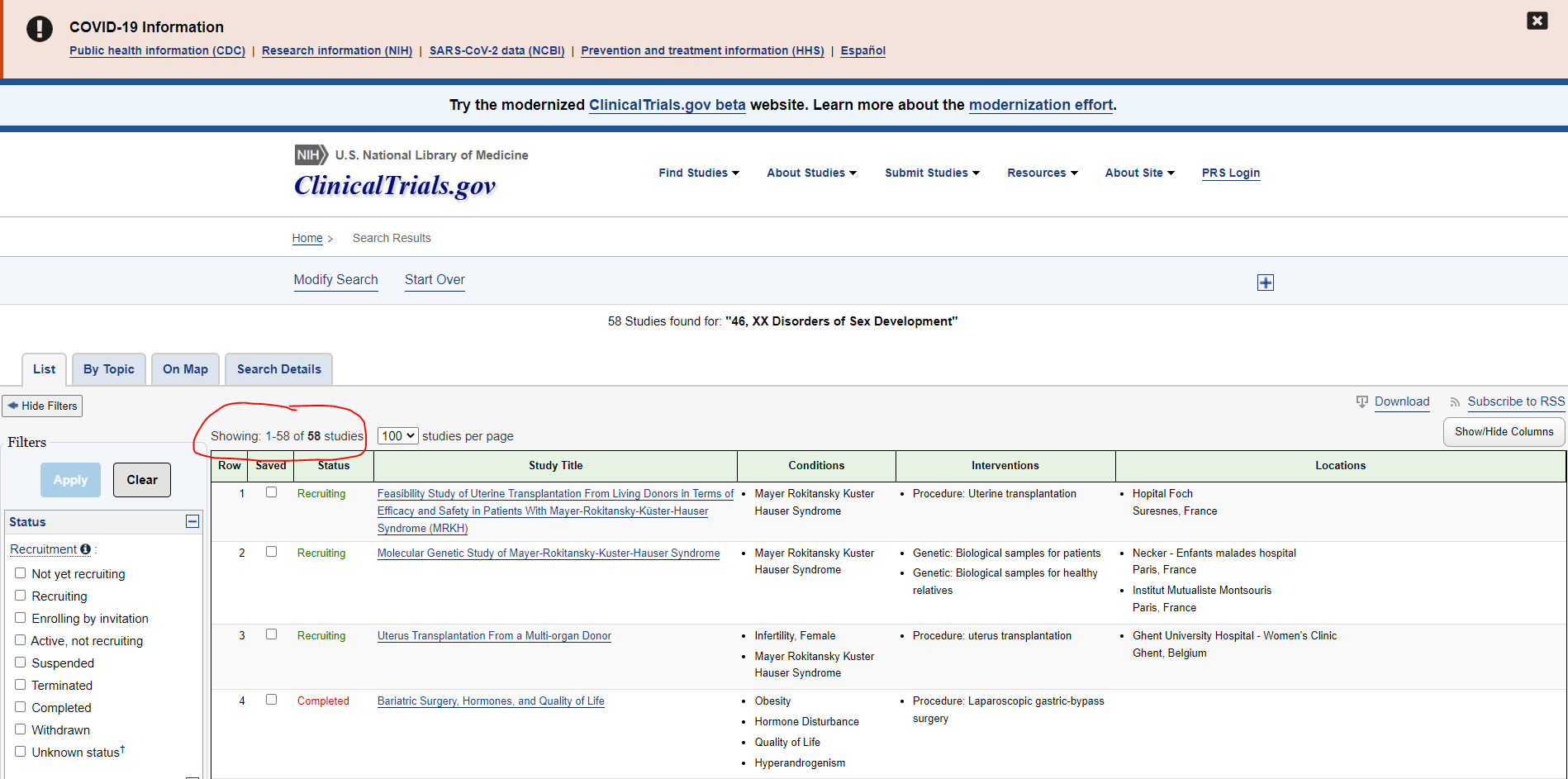
My goal is to extract data from clinicaltrials. On each page there is a table, I try to extract the data table from each page then concatenate all the tables into one. My code works but there are two problems, the first one is that sometimes (depending on the link) the data table has not the correct structure like the following
Row Saved Status Study Title Conditions Interventions Locations
0 1 NaN Completed Pilot Survey Arbovirus NaN NaN
1 Locations: Tropical Medicine centerKaohsiung c... Locations: Tropical Medicine centerKaohsiung c... Locations: Tropical Medicine centerKaohsiung c... Locations: Tropical Medicine centerKaohsiung c... Locations: Tropical Medicine centerKaohsiung c... Locations: Tropical Medicine centerKaohsiung c... NaN
as you can see the second row has the column Location repeated multiple times ..
here's my code :
import time
import selenium
from selenium import webdriver
from selenium.webdriver.common.by import By
from selenium.webdriver.support.ui import Select
from selenium.webdriver.chrome.service import Service
from selenium.webdriver.chrome.options import Options
from selenium.webdriver.support.ui import WebDriverWait
import html5lib
from bs4 import BeautifulSoup
import pandas as pd
from selenium.common.exceptions import TimeoutException
from selenium.common.exceptions import WebDriverException
from selenium.webdriver.support import expected_conditions as EC
ser = Service("/xxxxxxx/chromedriver") # change it to where your chromedriver is
options = webdriver.ChromeOptions()
options.add_argument("--headless")
options.add_argument("start-maximized")
options.add_argument('disable-infobars')
driver = webdriver.Chrome(service=ser, options=options)
#url = "https://clinicaltrials.gov/ct2/results?cond=Abruptio Placentae"
url = "https://www.clinicaltrials.gov/ct2/results?cond="46, XX Disorders of Sex Development""
driver.get(url)
#select = Select(driver.find_element_by_name('theDataTable_length'))
#select.select_by_value('100')
time.sleep(1)
html = driver.page_source
soup = BeautifulSoup(html, 'html.parser')
list_table = []
#df_tables = pd.read_html(str(soup), flavor="bs4")
df_tables = pd.read_html(str(soup))[1]
list_table.append(df_tables)
xp_next = '//*[@id="theDataTable_next"]/span'
while True:
try:
WebDriverWait(driver, 2).until(EC.visibility_of_element_located(
(By.CSS_SELECTOR, "[class='paginate_button next']")))
driver.find_element_by_xpath(xp_next).click()
html = driver.page_source
soup = BeautifulSoup(html, 'html.parser')
df_tables = pd.read_html(str(soup))[1]
#display(df_tables)
list_table.append(df_tables)
print("Navigating to Next Page")
time.sleep(1)
except (TimeoutException, WebDriverException) as e:
print("Last page reached")
break
driver.quit()
df = pd.concat(list_table)
df
I'm thinking maybe the problem is read_html?
CodePudding user response:
I think it may just be easer to go after the data directly. No need to use selenium then. Just need to parse some of the html:
import pandas as pd
import requests
from bs4 import BeautifulSoup
url = 'https://www.clinicaltrials.gov/ct2/results/rpc/mi0yqBc9u64Wpg4BvnGkBnjPewc9S6hHSwS3Z6p3C61JJPhHc67aZwhLzdUVp'
payload = '''draw=3&columns[0][data]=0&columns[0][name]=&columns[0][searchable]=true&columns[0][orderable]=false&columns[0][search][value]=&columns[0][search][regex]=false&columns[1][data]=1&columns[1][name]=&columns[1][searchable]=false&columns[1][orderable]=false&columns[1][search][value]=&columns[1][search][regex]=false&columns[2][data]=2&columns[2][name]=&columns[2][searchable]=true&columns[2][orderable]=false&columns[2][search][value]=&columns[2][search][regex]=false&columns[3][data]=3&columns[3][name]=&columns[3][searchable]=true&columns[3][orderable]=false&columns[3][search][value]=&columns[3][search][regex]=false&columns[4][data]=4&columns[4][name]=&columns[4][searchable]=true&columns[4][orderable]=false&columns[4][search][value]=&columns[4][search][regex]=false&columns[5][data]=5&columns[5][name]=&columns[5][searchable]=true&columns[5][orderable]=false&columns[5][search][value]=&columns[5][search][regex]=false&columns[6][data]=6&columns[6][name]=&columns[6][searchable]=true&columns[6][orderable]=false&columns[6][search][value]=&columns[6][search][regex]=false&columns[7][data]=7&columns[7][name]=&columns[7][searchable]=true&columns[7][orderable]=false&columns[7][search][value]=&columns[7][search][regex]=false&columns[8][data]=8&columns[8][name]=&columns[8][searchable]=true&columns[8][orderable]=false&columns[8][search][value]=&columns[8][search][regex]=false&columns[9][data]=9&columns[9][name]=&columns[9][searchable]=true&columns[9][orderable]=false&columns[9][search][value]=&columns[9][search][regex]=false&columns[10][data]=10&columns[10][name]=&columns[10][searchable]=true&columns[10][orderable]=false&columns[10][search][value]=&columns[10][search][regex]=false&columns[11][data]=11&columns[11][name]=&columns[11][searchable]=true&columns[11][orderable]=false&columns[11][search][value]=&columns[11][search][regex]=false&columns[12][data]=12&columns[12][name]=&columns[12][searchable]=true&columns[12][orderable]=false&columns[12][search][value]=&columns[12][search][regex]=false&columns[13][data]=13&columns[13][name]=&columns[13][searchable]=true&columns[13][orderable]=false&columns[13][search][value]=&columns[13][search][regex]=false&columns[14][data]=14&columns[14][name]=&columns[14][searchable]=true&columns[14][orderable]=false&columns[14][search][value]=&columns[14][search][regex]=false&columns[15][data]=15&columns[15][name]=&columns[15][searchable]=true&columns[15][orderable]=false&columns[15][search][value]=&columns[15][search][regex]=false&columns[16][data]=16&columns[16][name]=&columns[16][searchable]=true&columns[16][orderable]=false&columns[16][search][value]=&columns[16][search][regex]=false&columns[17][data]=17&columns[17][name]=&columns[17][searchable]=true&columns[17][orderable]=false&columns[17][search][value]=&columns[17][search][regex]=false&columns[18][data]=18&columns[18][name]=&columns[18][searchable]=true&columns[18][orderable]=false&columns[18][search][value]=&columns[18][search][regex]=false&columns[19][data]=19&columns[19][name]=&columns[19][searchable]=true&columns[19][orderable]=false&columns[19][search][value]=&columns[19][search][regex]=false&columns[20][data]=20&columns[20][name]=&columns[20][searchable]=true&columns[20][orderable]=false&columns[20][search][value]=&columns[20][search][regex]=false&columns[21][data]=21&columns[21][name]=&columns[21][searchable]=true&columns[21][orderable]=false&columns[21][search][value]=&columns[21][search][regex]=false&columns[22][data]=22&columns[22][name]=&columns[22][searchable]=true&columns[22][orderable]=false&columns[22][search][value]=&columns[22][search][regex]=false&columns[23][data]=23&columns[23][name]=&columns[23][searchable]=true&columns[23][orderable]=false&columns[23][search][value]=&columns[23][search][regex]=false&columns[24][data]=24&columns[24][name]=&columns[24][searchable]=true&columns[24][orderable]=false&columns[24][search][value]=&columns[24][search][regex]=false&columns[25][data]=25&columns[25][name]=&columns[25][searchable]=true&columns[25][orderable]=false&columns[25][search][value]=&columns[25][search][regex]=false&start=0&length=100&search[value]=&search[regex]=false'''
headers = {'user-agent': 'Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/96.0.4664.93 Safari/537.36'}
jsonData = requests.post(url, data=payload, headers=headers).json()
df = pd.DataFrame(jsonData['data'])
for col in df.columns:
df[col] = df[col].apply(lambda row: BeautifulSoup(row, 'html.parser').text.strip())
**Output:
print(df.head(5).to_string())
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25
0 1 NCT03689842 Recruiting Feasibility Study of Uterine Transplantation From Living Donors in Terms of Efficacy and Safety in Patients With Mayer-Rokitansky-Küster-Hauser Syndrome (MRKH) Mayer Rokitansky Kuster Hauser Syndrome Procedure: Uterine transplantation Interventional Not Applicable Hopital Foch Other Allocation: N/AIntervention Model: Single Group AssignmentMasking: None (Open Label)Primary Purpose: Treatment PregnancySafety assessment of the donor, the recipient and the fœtusPsychological assessment of the donor and the recipientGraft Rejection assessment 20 Female 18 Years to 65 Years (Adult, Older Adult) NCT03689842 2016/26 December 14, 2017 December 14, 2024 June 1, 2025 September 28, 2018 August 14, 2019 Hopital FochSuresnes, France
1 2 NCT02967822 Recruiting Molecular Genetic Study of Mayer-Rokitansky-Kuster-Hauser Syndrome Mayer Rokitansky Kuster Hauser Syndrome Genetic: Biological samples for patientsGenetic: Biological samples for healthy relatives Observational Imagine InstituteReference center for rare diseases (Rare Gynecologic Diseases) Other Observational Model: CohortTime Perspective: Prospective Number of identified nucleotidic variation(s) whose consequences can explain the phenotype of MRKH syndrome 410 All Child, Adult, Older Adult NCT02967822 IMNIS2015-06 MRKH May 2016 May 2031 May 2031 November 18, 2016 October 12, 2018 Necker - Enfants malades hospitalParis, FranceInstitut Mutualiste MontsourisParis, France
2 3 NCT03252795 Recruiting Uterus Transplantation From a Multi-organ Donor Infertility, FemaleMayer Rokitansky Kuster Hauser Syndrome Procedure: uterus transplantation Interventional Not Applicable University Hospital, Ghent Other Allocation: N/AIntervention Model: Single Group AssignmentMasking: None (Open Label)Primary Purpose: Treatment Survival of the uterus 1 year after transplantationComplications after uterus transplantationOngoing pregnancy rateTake home baby rate 20 Female 18 Years to 38 Years (Adult) NCT03252795 EC/2016/0731 November 3, 2016 December 2021 December 2023 August 17, 2017 November 13, 2019 Ghent University Hospital - Women's ClinicGhent, Belgium
3 4 NCT03188640 Completed Bariatric Surgery, Hormones, and Quality of Life ObesityHormone DisturbanceQuality of LifeHyperandrogenism Procedure: Laparoscopic gastric-bypass surgery Interventional Not Applicable Linkoeping University Other Allocation: N/AIntervention Model: Single Group AssignmentMasking: None (Open Label)Primary Purpose: Treatment Sex-hormone levelsFemale sexual functionHormone-related quality of life(and 2 more...) 68 Female 18 Years to 50 Years (Adult) NCT03188640 2012/392-31 OBLIV March 1, 2014 October 31, 2016 October 31, 2016 June 15, 2017 June 16, 2017
4 5 NCT04912648 Not yet recruiting FEmale Metabolic Risk and Androgens: an Irish Longitudinal (FEMAIL) Study HyperandrogenismMetabolic DiseaseSex Hormones Adverse Reaction Other: Longitudinal follow up Observational Royal College of Surgeons, Ireland Other Observational Model: CohortTime Perspective: Prospective Correlation of sex hormone profiles and the metabolome of women across the lifespan with risk of metabolic dysfunction, cardiovascular disease and quality of life.Development of risk prediction models for development of diabetes in womenEstablishment of the longitudinal FEMAIL study database 500 Female 18 Years and older (Adult, Older Adult) NCT04912648 20/49 FEMAIL September 1, 2021 September 1, 2025 August 31, 2031 June 3, 2021 June 3, 2021 Royal College of Surgeons in IrelandDublin, Ireland
[58 rows x 26 columns]
CodePudding user response:
What happens?
Issue is caused in headless mode in combination with your arguments - Tables structure is swapping in responsive mode and some columns changes to rows.
How to fix?
You have to set options more strict and can force webdriver to use a specific window-size while start-maximized
options = webdriver.ChromeOptions()
options.add_argument("--window-size=1920,1080")
options.add_argument("--start-maximized")
options.add_argument("--headless")
Example
import selenium, html5lib, time
import pandas as pd
from bs4 import BeautifulSoup
from selenium import webdriver
from selenium.webdriver.common.by import By
from selenium.webdriver.support.ui import Select
from selenium.webdriver.chrome.service import Service
from selenium.webdriver.chrome.options import Options
from selenium.webdriver.support.ui import WebDriverWait
from selenium.common.exceptions import TimeoutException
from selenium.common.exceptions import WebDriverException
from selenium.webdriver.support import expected_conditions as EC
ser = Service("#####################") # change it to where your chromedriver is
options = webdriver.ChromeOptions()
options.add_argument("--window-size=1920,1080")
options.add_argument("--start-maximized")
options.add_argument("--headless")
driver = webdriver.Chrome(service=ser, options=options)
url = "https://www.clinicaltrials.gov/ct2/results?cond="46, XX Disorders of Sex Development""
driver.get(url)
list_table = []
while True:
time.sleep(1)
html = driver.page_source
soup = BeautifulSoup(html, 'html.parser')
list_table.append(pd.read_html(html)[1])
print("Navigating to Next Page")
try:
WebDriverWait(driver, 10).until(EC.element_to_be_clickable((By.CSS_SELECTOR, "[class='paginate_button next'] span"))).click()
except (TimeoutException, WebDriverException) as e:
print("Last page reached")
break
driver.quit()
df = pd.concat(list_table)
df
Outout
| Row | Saved | Status | Study Title | Conditions | Interventions | Locations | |
|---|---|---|---|---|---|---|---|
| 0 | 1 | nan | Recruiting | Feasibility Study of Uterine Transplantation From Living Donors in Terms of Efficacy and Safety in Patients With Mayer-Rokitansky-Küster-Hauser Syndrome (MRKH) | Mayer Rokitansky Kuster Hauser Syndrome | Procedure: Uterine transplantation | Hopital FochSuresnes, France |
| 1 | 2 | nan | Recruiting | Molecular Genetic Study of Mayer-Rokitansky-Kuster-Hauser Syndrome | Mayer Rokitansky Kuster Hauser Syndrome | Genetic: Biological samples for patientsGenetic: Biological samples for healthy relatives | Necker - Enfants malades hospitalParis, FranceInstitut Mutualiste MontsourisParis, France |
| 2 | 3 | nan | Recruiting | Uterus Transplantation From a Multi-organ Donor | Infertility, FemaleMayer Rokitansky Kuster Hauser Syndrome | Procedure: uterus transplantation | Ghent University Hospital - Women's ClinicGhent, Belgium |