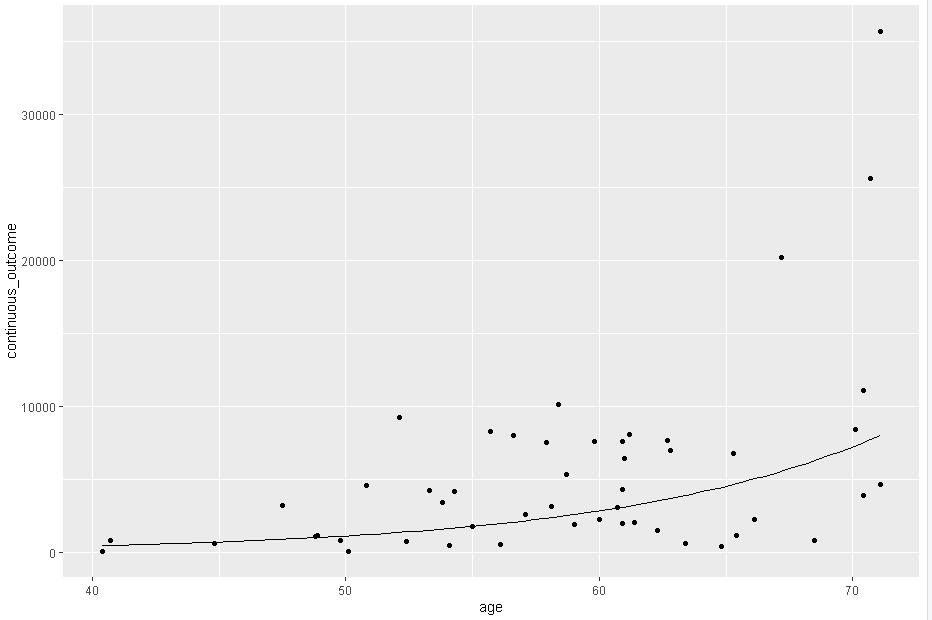
To make the results that I obtained from a linear mixed model more insightful, I'm trying to plot the predicted values with a 95% confidence interval.
The data and the model that I specified look as follows:
library(ggplot2)
library(magrittr)
library(nlme)
mydata <- structure(list(pat_id = c(61, 8, 179, 13, 153, 80, 273, 142,
202, 2, 61, 127, 135, 49, 20, 25, 315, 48, 157, 171, 29, 271,
105, 14, 14, 84, 180, 125, 255, 135, 39, 49, 320, 160, 197, 189,
154, 77, 30, 75, 50, 103, 115, 80, 88, 34, 310, 81, 75, 82),
age = c(57.1, 61.4, 60.9, 58.7, 65.4, 44.8, 59, 47.5, 63.4,
59.8, 55, 66.1, 50.8, 62.8, 71.1, 48.9, 58.1, 49.8, 71.1,
56.6, 50.1, 40.4, 62.3, 53.3, 53.8, 70.4, 67.2, 68.5, 57.9,
54.3, 60.9, 61, 54.1, 70.1, 52.4, 70.4, 55.7, 60.7, 56.1,
60.9, 60, 52.1, 48.8, 40.7, 58.4, 65.3, 62.7, 64.8, 61.2,
70.7), continuous_outcome = c(2605.440900389, 2063.9, 1929.46472167969,
5341.496223986, 1119.6044921885, 566.255950927734, 1880.5290222168,
3215.77453613281, 603.7692803513, 7594.527875667, 1775.66680908203,
2260.04156843471, 4565.098635412, 6948.76321012, 35685.6,
1170.96138000488, 3152.85720825195, 765.9703779527, 4638.270794249,
7962.890625, 45.4402923583984, 76.2939453125, 1466.9, 4207.615392583,
3410.15625, 11076.9451141357, 20190.12993463, 822.127334531,
7512.78955078125, 4146.38368225098, 4292.35992431641, 6393.09959411621,
465.39306640625, 8409, 737.1825748757, 3898.624597663, 8238.67454528809,
3054.98580932617, 526.4287617193, 7612.90808598633, 2247.54676818848,
9231.37016296387, 1067.416015625, 775.980377197266, 10118.2,
6744.3, 7616.49208068848, 356.531524658203, 8069.160616877,
25596.8906372702), continuous_outcome_log = c(7.86574093001633,
7.63283707807243, 7.56551604134912, 8.58344828040522, 7.0216235438582,
6.34081061444271, 7.53984003495667, 8.07613443965171, 6.40484703050537,
8.93531491566494, 7.48249430865052, 7.72358085708673, 8.42641442900534,
8.84646286749209, 10.4825305471662, 7.06643401737976, 8.05638149327426,
6.64244817996005, 8.44231247645808, 8.98267293141324, 3.83816745221036,
4.34761562539111, 7.29158808696319, 8.34488898720956, 8.13480658905054,
9.31271148408861, 9.9129986748043, 6.71311090867393, 8.92449521812674,
8.33023297640332, 8.36482490452301, 8.76313090568181, 6.14502876872992,
9.03717675296699, 6.60419118543475, 8.26863557047525, 9.01671612519243,
8.02485750673053, 6.26801380735694, 8.93773186515778, 7.71803940572248,
9.13047108358574, 6.97393247141267, 6.65541509547637, 9.22218988833333,
8.81660124504343, 8.93820247123851, 5.87922353910847, 8.99592866402467,
10.1502652300912), fu_time = c(2.19, 4.005, 0, 0.857, 0.745,
4.153, 1.522, 0, 0.12, 3.184, 0, 0.802, 0.517, 1.859, 0.268,
0, 0, 0.799, 0.446, 0.78, 0, 0, 2.185, 0.953, 1.457, 3.888,
0.268, 1.295, 0, 3.967, 4.375, 0, 0, 0.479, 0.159, 1.29,
0, 0, 0.734, 0, 0, 0, 3.584, 0, 0.997, 1.106, 1.506, 0, 0.307,
0)), row.names = c(NA, -50L), class = c("tbl_df", "tbl",
"data.frame"))
regression <-
lme(fixed=continuous_outcome_log ~ 1 fu_time age*fu_time,
random=~1|pat_id,
data=mydata,
correlation=corCAR1(form=~1 fu_time|pat_id),
na.action="na.omit",
method="REML",
control=lmeControl(opt="optim"))
To plot the predicted values I've first created a prediction dataframe (pframe) as follows:
pframe <-
expand.grid(
fu_time=mean(mydata$fu_time),
age=seq(min(mydata$age), max(mydata$age), length.out=75))
pframe$predicted_continuous_outcome <-
exp(predict(regression, newdata=pframe, level=0))
Next, I've plotted the predicted values over the observed values as follows:
predict_plot <-
mydata %>%
ggplot(
data=.,
aes(x=age, y=continuous_outcome))
geom_point()
geom_line(
data=pframe,
aes(x=age, y=predicted_continuous_outcome))
predict_plot
How do I predict and plot a 95% confidence interval around the predictions in pframe? I tried pframe$ci <- exp(predict(regression, newdata=pframe, interval="confidence")) but that produces an error: Error in predict.lme(regression, newdata = pframe, interval = "confidence") :
cannot evaluate groups for desired levels on 'newdata'
CodePudding user response:
There doesn't appear to be a predict method for lme that can do confidence intervals. But they can be calculated manually (following calculations here). Possibly a good job for a function to predict your three values of interest:
library(ggplot2)
library(magrittr)
library(nlme)
library(dplyr)
pframe <-
expand.grid(
fu_time=mean(mydata$fu_time),
age=seq(min(mydata$age), max(mydata$age), length.out=75))
constructCIRibbon <- function(newdata, model) {
df <- newdata %>%
mutate(Predict = predict(model, newdata = ., level = 0))
mm <- model.matrix(eval(eval(model$call$fixed)[-2]), data = df)
vars <- mm %*% vcov(model) %*% t(mm)
sds <- sqrt(diag(vars))
df %>% mutate(
lowCI = Predict - 1.96 * sds,
HiCI = Predict 1.96 * sds,
across(Predict:HiCI, exp)
)
}
pframe <- constructCIRibbon(pframe, regression)
mydata %>%
ggplot(aes(age, continuous_outcome))
geom_point()
geom_line(data = pframe, aes(y = Predict))
geom_ribbon(data = pframe,
aes(y = Predict, ymin = lowCI, ymax = HiCI),
alpha = 0.2)
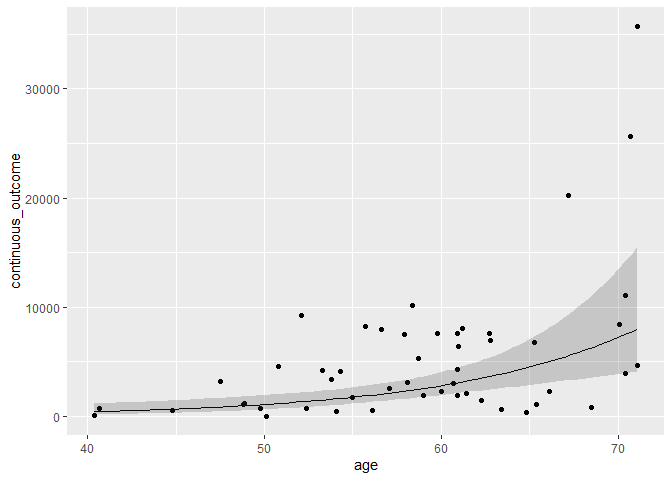
Created on 2021-12-15 by the reprex package (v2.0.1)