I have a problem with rewrite variable in reactive output. In my code df1() is uploaded main dataset. In modified_df1 i changed data type of variable based on user select. My problem is, in final dataset modified_df1 a I get only new column var (because I use cbind(df1(), var)). Is there some way, how I can rewrite previous column, no add new with cbind()? I pase server code here, thanks.
df1 <- reactive({
req(input$file1)
df <- read.csv(input$file1$datapath,
header = input$header,
sep = input$sep,
quote = input$quote)
df
})
colnames <- reactive({ names(df1()) })
observeEvent(df1(), {
updateCheckboxGroupInput(session, "class_var",
label = "Select Columns",
choices = colnames(),
selected = "")
})
## update when selection changes
# storing the selected variable from the variables list table
table.sel <- reactive({
df1()[,which(colnames(df1()) == col.name()[input$class_var,1])]
})
modified_df1 = eventReactive(input$chg_class,{
if( input$choose_class == "Numeric"){
var <- as.numeric(df1()[, input$class_var])
} else if(input$choose_class == "Factor"){ message("get new vars");
var <- as.factor(df1()[, input$class_var])
} else if( input$choose_class == "Character"){
var <- as.character(df1()[, input$class_var])
} else if( input$choose_class == "Date"){
var <- as.Date(df1()[, input$class_var])
}
df2 = cbind(df1(), var)
})
CodePudding user response:
Since we are selecting multiple columns, we can use dplyr::across to loop through them. For example if you want to change the columns selected to a character:
var <- df1() %>% mutate(across(all_of(input$class_var), ~as.character(.)))
I made an app to ilustrate better what's happening.
library(shiny)
library(tidyverse)
write_csv(iris, file = "iris.csv")
# ui ----------------------------------------------------------------------
ui <- fluidPage(
fluidRow(
column(
width = 6,
fileInput(
"file1",
"Upload File",
multiple = FALSE,
accept = NULL,
width = NULL,
buttonLabel = "Browse...",
placeholder = "No file selected"
),
checkboxGroupInput("class_var", "Select Columns", choices = "", selected = ""),
radioButtons("choose_class", "Select Columns", choices = c("Numeric", "Factor", "Character", "Date")),
actionButton("chg_class", "Change Class")
),
column(
width = 6,
tableOutput("table_df")
)
)
)
# server ------------------------------------------------------------------
server <- function(input, output, session) {
df1 <- reactive({
req(input$file1)
read.csv(input$file1$datapath)
})
colnames <- reactive({
names(df1())
})
observeEvent(df1(), {
updateCheckboxGroupInput(session,
"class_var",
label = "Select Columns",
choices = colnames(),
selected = ""
)
})
## update when selection changes
# storing the selected variable from the variables list table
table.sel <- reactive({
df1()[, which(colnames(df1()) == col.name()[input$class_var, 1])]
})
modified_df1 <- eventReactive(input$chg_class, {
if (input$choose_class == "Numeric") {
var <- df1() %>% mutate(across(all_of(input$class_var), ~ as.numeric(.)))
} else if (input$choose_class == "Factor") {
message("get new vars")
var <- df1() %>% mutate(across(all_of(input$class_var), ~ as.factor(.)))
} else if (input$choose_class == "Character") {
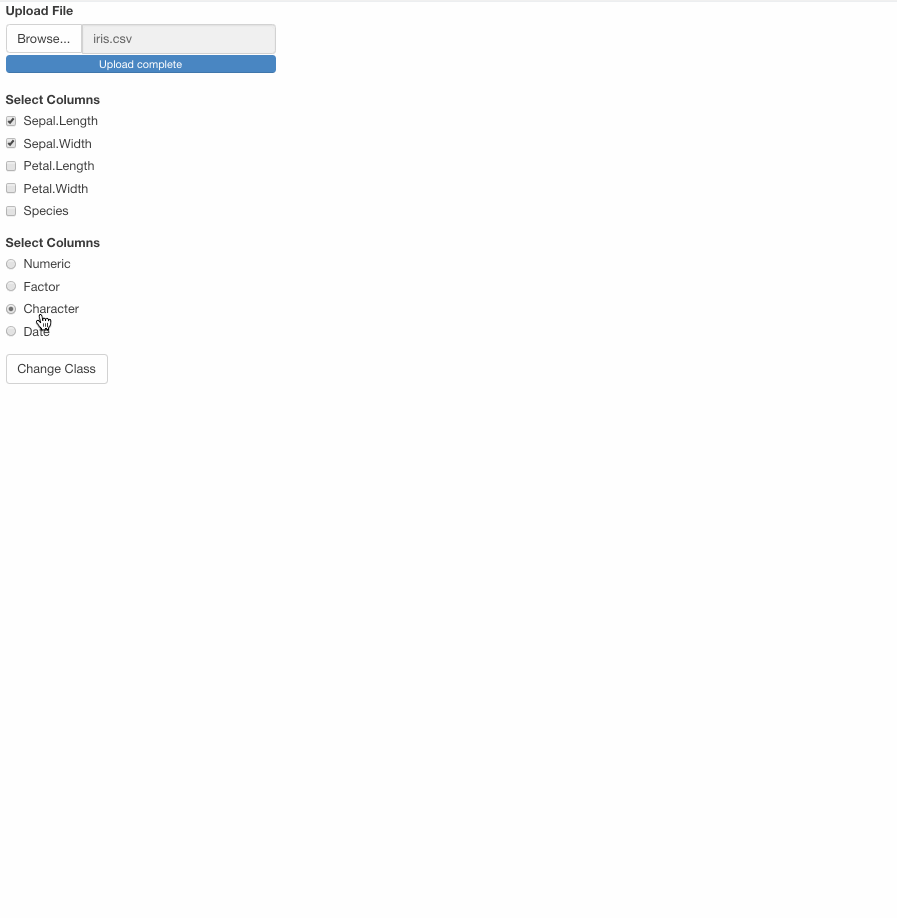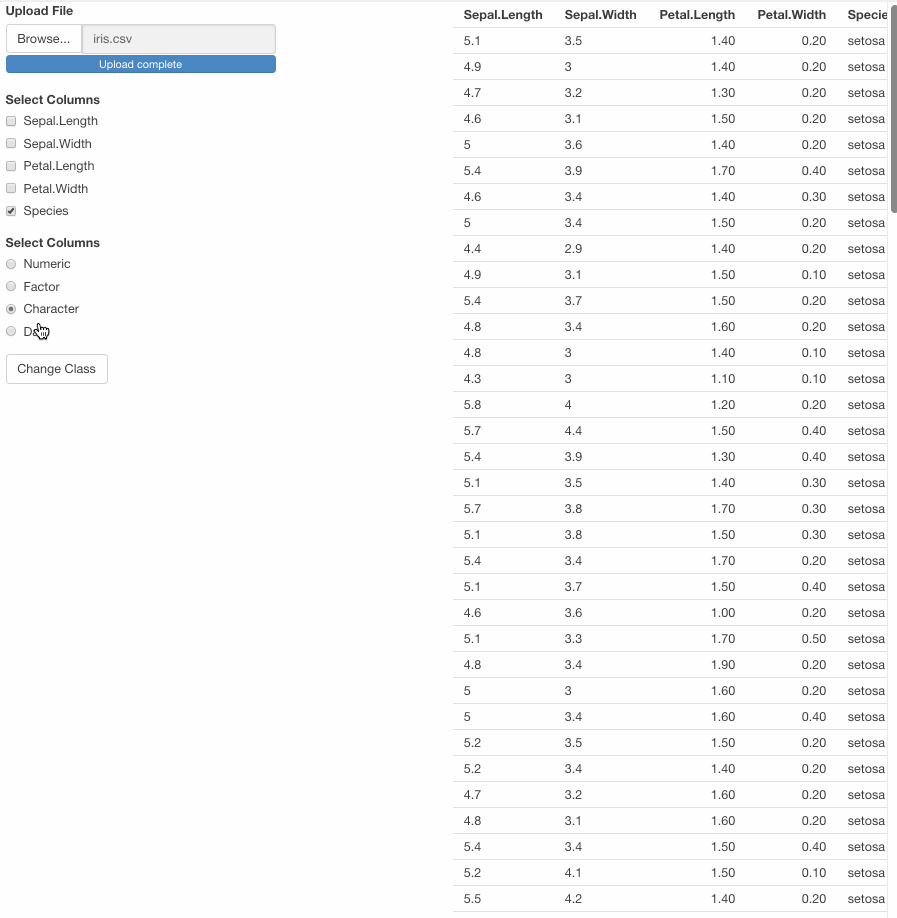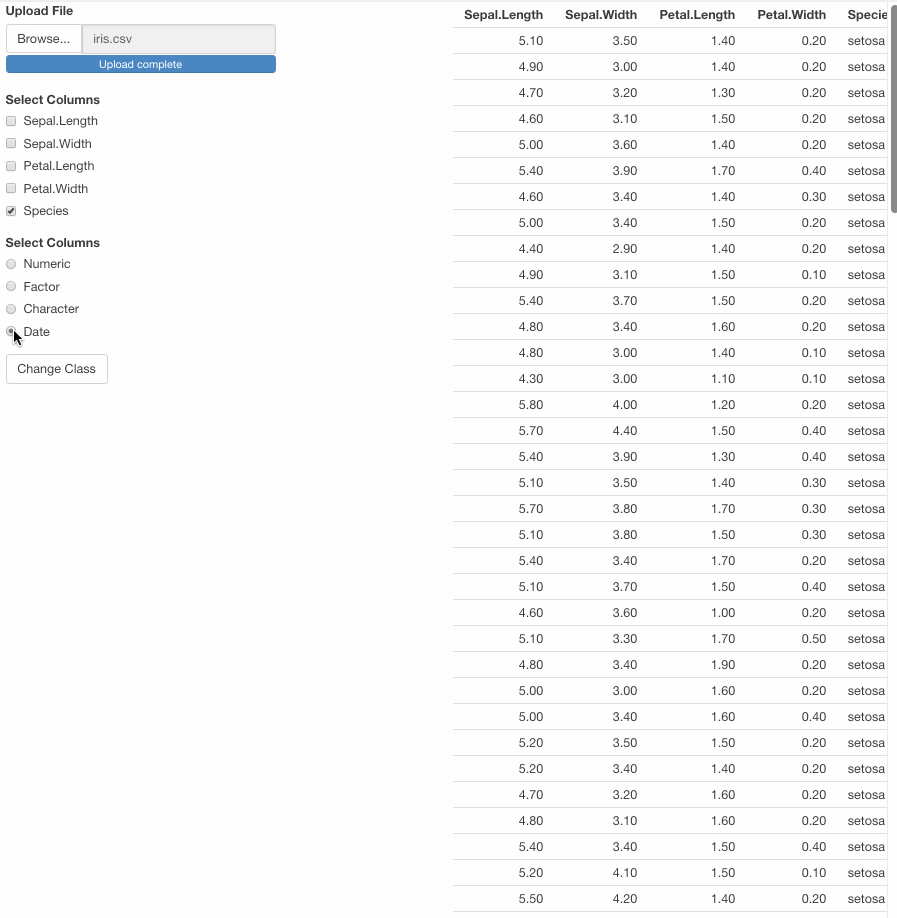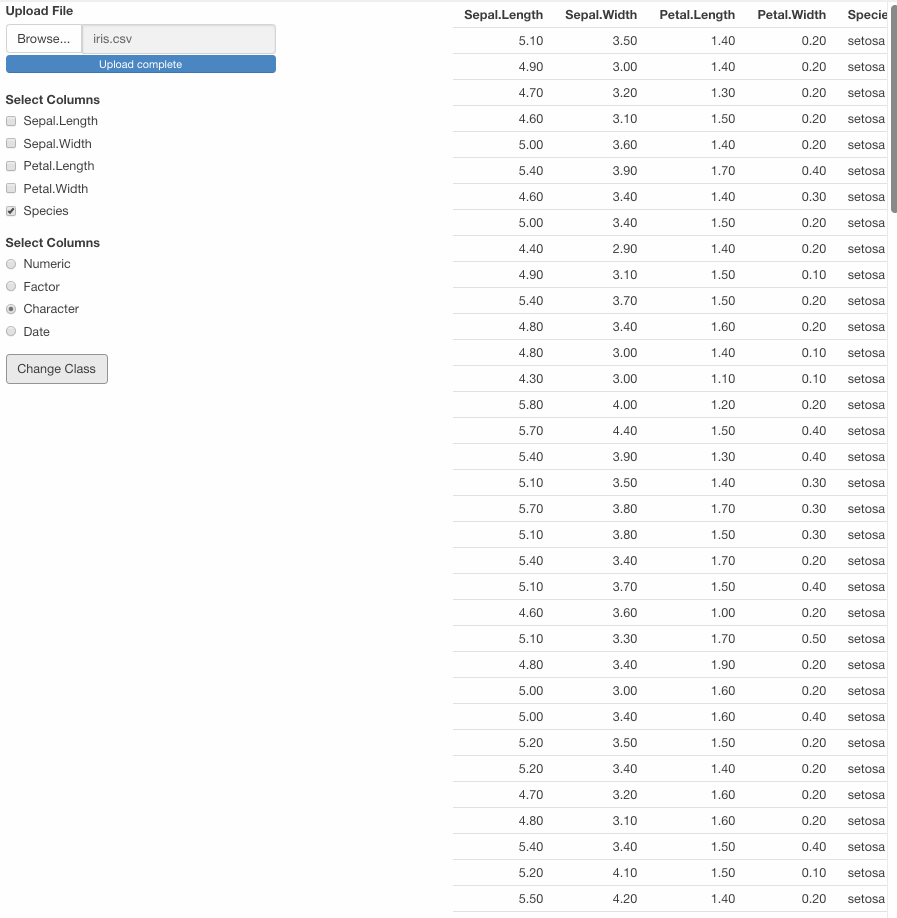
var <- df1() %>% mutate(across(all_of(input$class_var), ~ as.character(.)))
} else if (input$choose_class == "Date") {
var <- df1() %>% mutate(across(all_of(input$class_var), ~ as.Date(.)))
}
var
})
output$table_df <- renderTable({
modified_df1()
})
}
shinyApp(ui, server)
Note that if you try to convert to Date a character vector an error will occur.