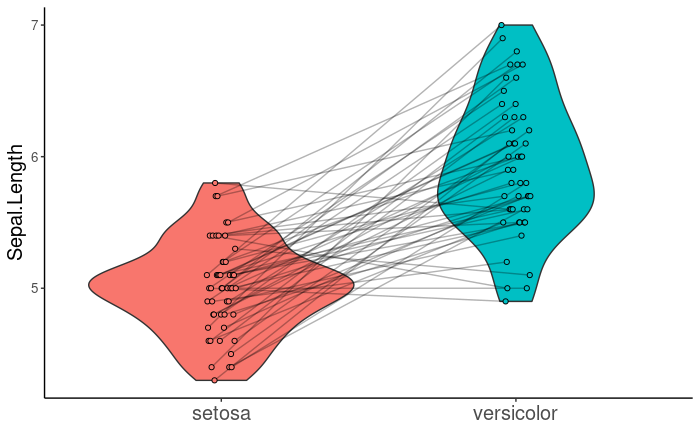
With ggplot2, I can create a violin plot with overlapping points, and paired points can be connected using geom_line().
library(datasets)
library(ggplot2)
library(dplyr)
iris_edit <- iris %>% group_by(Species) %>%
mutate(paired = seq(1:length(Species))) %>%
filter(Species %in% c("setosa","versicolor"))
ggplot(data = iris_edit,
mapping = aes(x = Species, y = Sepal.Length, fill = Species))
geom_violin()
geom_line(mapping = aes(group = paired),
position = position_dodge(0.1),
alpha = 0.3)
geom_point(mapping = aes(fill = Species, group = paired),
size = 1.5, shape = 21,
position = position_dodge(0.1))
theme_classic()
theme(legend.position = "none",
axis.text.x = element_text(size = 15),
axis.title.y = element_text(size = 15),
axis.title.x = element_blank(),
axis.text.y = element_text(size = 10))
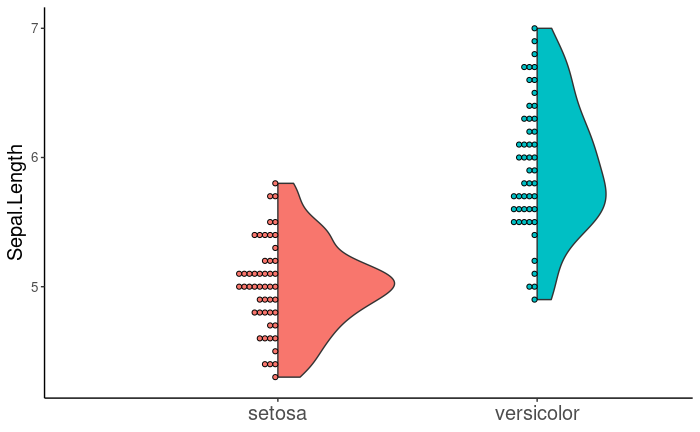
The see package includes the geom_violindot() function to plot a halved violin plot alongside its constituent points. I've found this function helpful when plotting a large number of points so that the violin is not obscured.
library(see)
ggplot(data = iris_edit,
mapping = aes(x = Species, y = Sepal.Length, fill = Species))
geom_violindot(dots_size = 0.8,
position_dots = position_dodge(0.1))
theme_classic()
theme(legend.position = "none",
axis.text.x = element_text(size = 15),
axis.title.y = element_text(size = 15),
axis.title.x = element_blank(),
axis.text.y = element_text(size = 10))
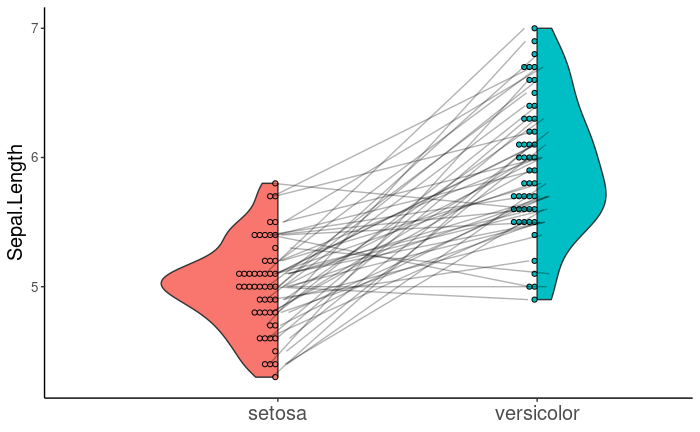
Now, I would like to add geom_line() to geom_violindot() in order to connect paired points, as in the first image. Ideally, I would like the points to be inside and the violins to be outside so that the lines do not intersect the violins. geom_violindot() includes the flip argument, which takes a numeric vector specifying the geoms to be flipped.
ggplot(data = iris_edit,
mapping = aes(x = Species, y = Sepal.Length, fill = Species))
geom_violindot(dots_size = 0.8,
position_dots = position_dodge(0.1),
flip = c(1))
geom_line(mapping = aes(group = paired),
alpha = 0.3,
position = position_dodge(0.1))
theme_classic()
theme(legend.position = "none",
axis.text.x = element_text(size = 15),
axis.title.y = element_text(size = 15),
axis.title.x = element_blank(),
axis.text.y = element_text(size = 10))
As you can see, invoking flip inverts the violin half, but not the corresponding points. The see documentation does not seem to address this.
Questions
- How can you create a
geom_violindot()plot with paired points, such that the points and the lines connecting them are "sandwiched" in between the violin halves? I suspect there is a solution that uses David Robinson'sGeomFlatViolinfunction, though I haven't been able to figure it out. - In the last figure, note that the lines are askew relative to the points they connect. What position adjustment function should be supplied to the
position_dotsandpositionarguments so that the points and lines are properly aligned?
CodePudding user response:
Not sure about using geom_violindot with see package. But you could use a combo of geom_half_violon and geom_half_dotplot with gghalves package and subsetting the data to specify the orientation:
library(gghalves)
ggplot(data = iris_edit[iris_edit$Species == "setosa",],
mapping = aes(x = Species, y = Sepal.Length, fill = Species))
geom_half_violin(side = "l")
geom_half_dotplot(stackdir = "up")
geom_half_violin(data = iris_edit[iris_edit$Species == "versicolor",],
aes(x = Species, y = Sepal.Length, fill = Species), side = "r")
geom_half_dotplot(data = iris_edit[iris_edit$Species == "versicolor",],
aes(x = Species, y = Sepal.Length, fill = Species),stackdir = "down")
geom_line(data = iris_edit, mapping = aes(group = paired),
alpha = 0.3)
As a note, the lines in the pairing won't properly align because the dotplot is binning each observation then lengthing out the dotline-- the paired lines only correspond to x-value as defined in aes, not where the dot is in the line.
CodePudding user response:
As per comment - this is not a direct answer to your question, but I believe that you might not get the most convincing visualisation when using the "slope graph" optic. This becomes quickly convoluted (so many dots/ lines overlapping) and the message gets lost.
To show change between paired observations (treatment 1 versus treatment 2), you can also (and I think: better) use a scatter plot. You can show each observation and the change becomes immediately clear. To make it more intuitive, you can add a line of equality.
I don't think you need to show the estimated distribution (left plot), but if you want to show this, you could make use of a two-dimensional density estimation, with geom_density2d (right plot)
library(tidyverse)
## patchwork only for demo purpose
library(patchwork)
iris_edit <- iris %>% group_by(Species) %>%
## use seq_along instead
mutate(paired = seq_along(Species)) %>%
filter(Species %in% c("setosa","versicolor")) %>%
## some more modificiations
select(paired, Species, Sepal.Length) %>%
pivot_wider(names_from = Species, values_from = Sepal.Length)
lims <- c(0, 10)
p1 <-
ggplot(data = iris_edit, aes(setosa, versicolor))
geom_abline(intercept = 0, slope = 1, lty = 2)
geom_point(alpha = .7, stroke = 0, size = 2)
cowplot::theme_minimal_grid()
coord_equal(xlim = lims, ylim = lims)
labs(x = "Treatment 1", y = "Treatment 2")
p2 <-
ggplot(data = iris_edit, aes(setosa, versicolor))
geom_abline(intercept = 0, slope = 1, lty = 2)
geom_density2d(color = "Grey")
geom_point(alpha = .7, stroke = 0, size = 2)
cowplot::theme_minimal_grid()
coord_equal(xlim = lims, ylim = lims)
labs(x = "Treatment 1", y = "Treatment 2")
p1 p2
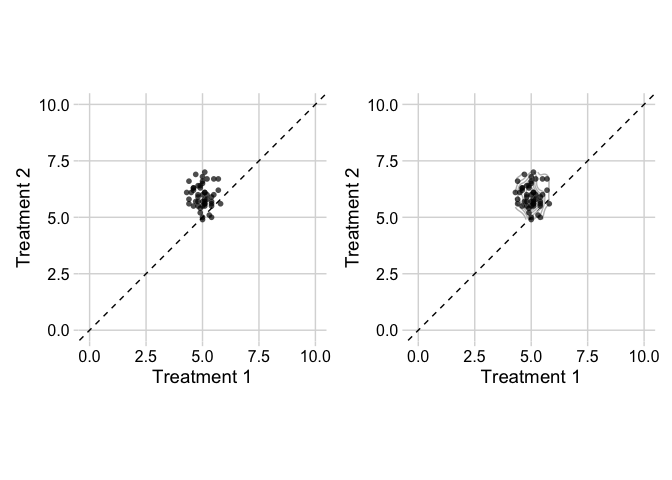
Created on 2021-12-18 by the reprex package (v2.0.1)