I am trying to create an app where you can create boxplots and add the Kruskal-Wallis p-value if the user chooses it.
The app has 3 tabs:
- Tab 1 > it has a
checkboxInputthat if you click on it, you will do the log2 transformation. In addition, it has oneactionButtonto submit your data (with or without log2). If you don't click the button, you won't be able to draw the plot. - Tab 2 > it has
radioButtonsandcheckboxInputwhich allows you draw different plots depending on the user's choice. - Tab 3 > it has a
numericInputthat allows you to change the opacity of the plot. In addition, it has acheckboxInputwhich allows you to add the Kruskal-Wallis p-value to the plot.
It works perfectly. However, when I want to add the KW p-value, the value changes by itself before clicking the actionButton.
This is how it looks when you have selected 2 groups and you have clicked the checkboxInput from tab3 to show the KW pvalue.
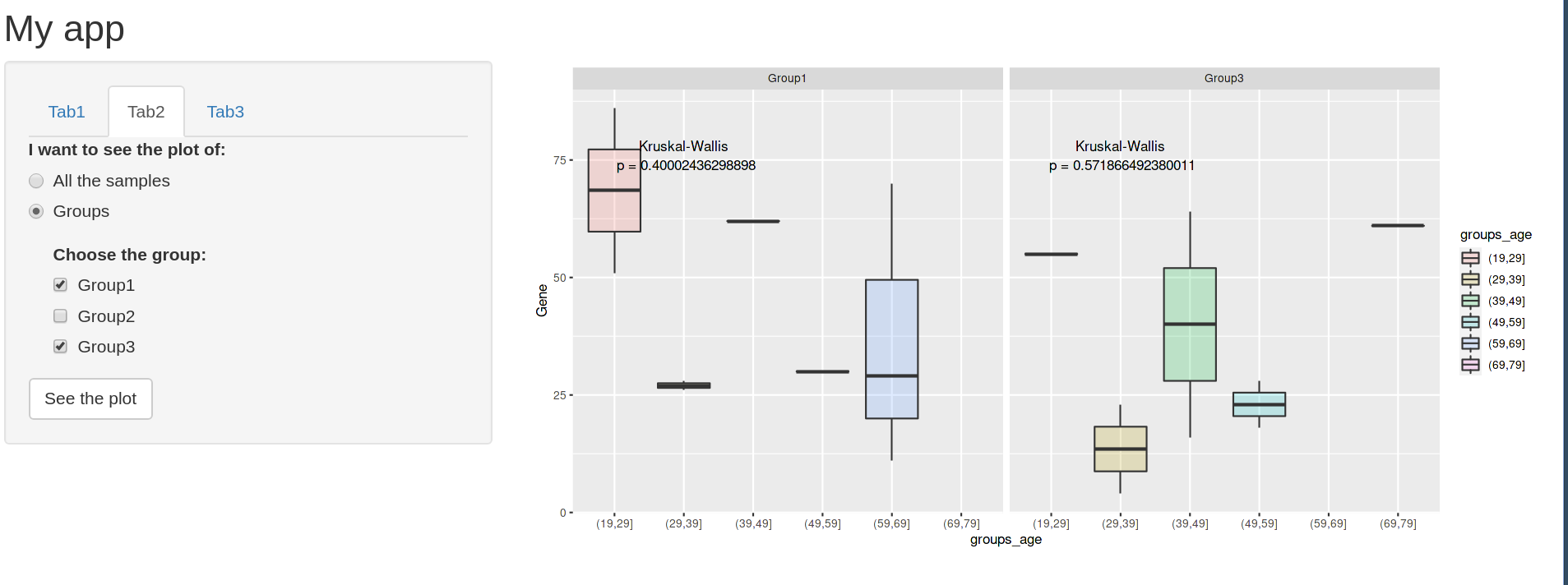
However, if you deselect the group 1, in order to only see Group 3, the place of the p-value changes before clicking the actionButton.
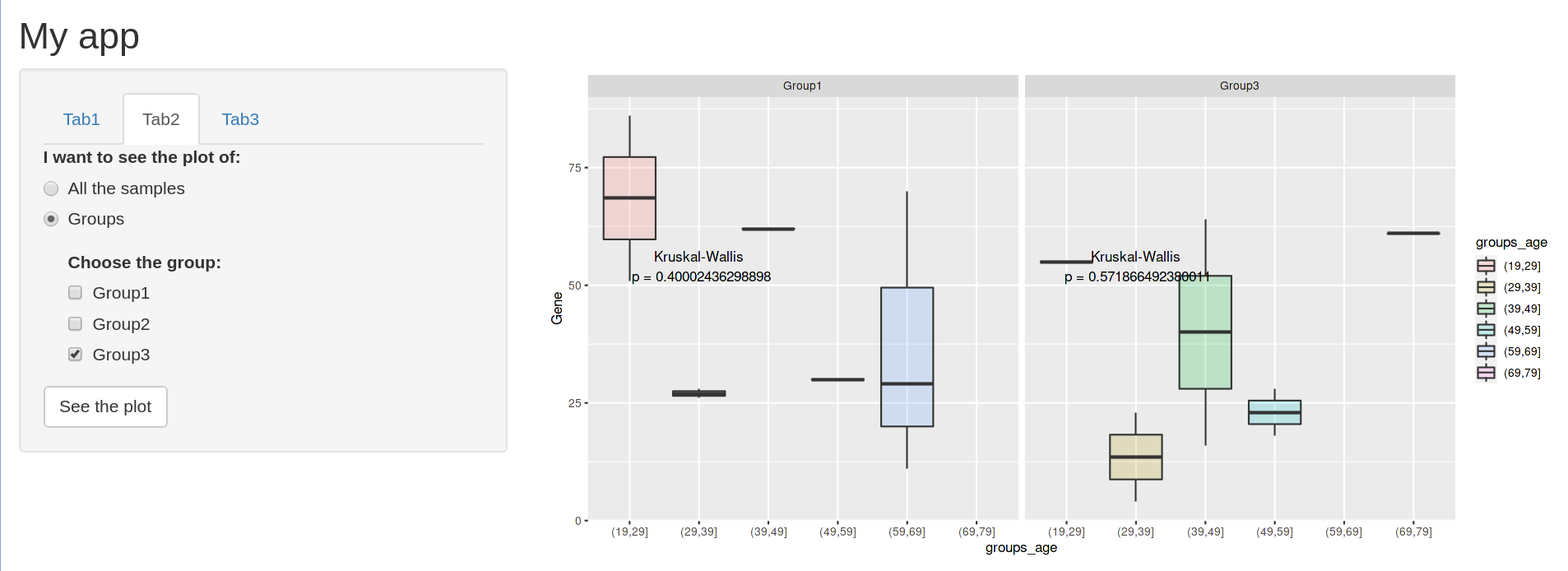
And then, when you click the button, you have the final output that you were expecting to have.
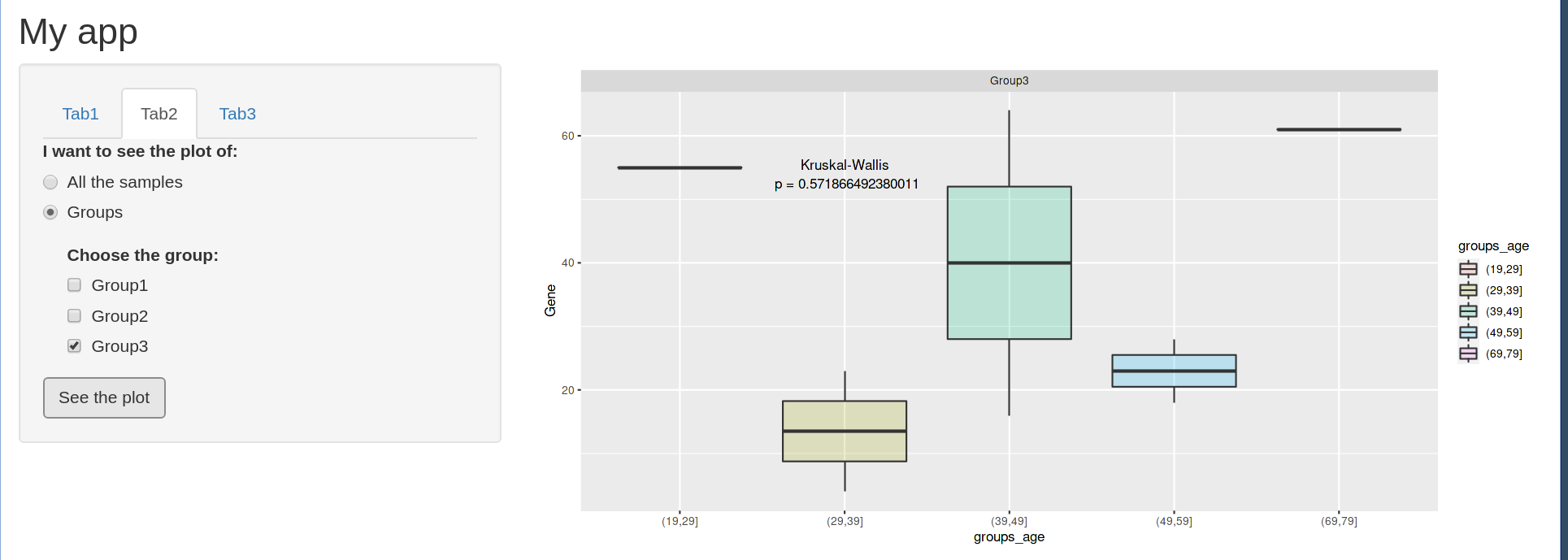
On the other hand, if the user decides to change the place of the p-value (through the numericInputs that they appear after clicking "Show the Kruskal Wallis p-value"), the plot updates without having the change to click the actionButton.
In conclusion, the problem is that the plot updates before clicking the actionButton and I don't know how to solve it.
Note that if you change the opacity of the plot, the plot won't change unless you click the actionButton (something that I want for all the app).
Does anyone know how to fix it?
Thanks in advance
The code:
library(shiny)
library(dplyr)
library(ggplot2)
ui <- fluidPage(
titlePanel("My app"),
sidebarLayout(
sidebarPanel(
tabsetPanel(
tabPanel("Tab1",
checkboxInput("log2", "Log2 transformation", value = FALSE),
actionButton("submit", "Submit")
),
tabPanel("Tab2",
radioButtons(inputId = "plot_type", label = "I want to see the plot of:",
c("All the samples" = "all_samples",
"Groups" = "samples_group")),
conditionalPanel(
condition = "input.plot_type == 'samples_group'",
style = "margin-left: 20px;",
checkboxGroupInput("group", "Choose the group:",
choices = c("Group1", "Group2", "Group3"))),
actionButton("show_plot", "See the plot")
),
tabPanel("Tab3",
numericInput("alpha", "Opacity of the plot", value=0.2),
checkboxInput(inputId = "Kruskalpval", label = "Show the Kruskal Wallis p-value", value = FALSE),
conditionalPanel(
condition = "input.Kruskalpval == '1'",
style = "margin-left: 20px;",
checkboxInput(inputId = "changeKW", "I want to change the place of the value", value=FALSE),
conditionalPanel(
condition = "input.changeKW == '1'",
numericInput(inputId = "X_axis", "X_axis:", value=2),
numericInput(inputId = "Y_axis", "Y_axis:", value=70)
)
),
actionButton("show_plot_2", "See the plot")
)
)
),
mainPanel(
plotOutput("boxplots")
)
)
)
server <- function(input, output) {
set.seed(1234)
Gene <- floor(runif(25, min=0, max=101))
groups_age <- floor(runif(25, min=18, max=75))
Group <- c("Group1", "Group1", "Group3", "Group2", "Group1", "Group3", "Group2", "Group2", "Group2", "Group1", "Group1", "Group3", "Group1", "Group2", "Group1", "Group2", "Group3", "Group1", "Group3", "Group3", "Group2", "Group1", "Group3", "Group3","Group2")
data <- reactive({
df <- data.frame(Gene, Group, groups_age)
mybreaks <- seq(min(df$groups_age)-1, to=max(df$groups_age) 10, by=10)
df$groups_age <- cut(df$groups_age, breaks = mybreaks, by=10)
if(input$plot_type == "samples_group"){
# if the user selects everything, it will take everything.
if(all(c("Group1", "Group2", "Group3") %in% input$group)){
return(df)
# if the user only selects group1 and group2, it will appear only those columns.
}else if (all(c("Group1", "Group2") %in% input$group)) {
df <- subset(df, (df$Group == "Group1" | df$Group == "Group2"))
return(df)
# if the user only selects group1 and group3, it will appear only those columns.
}else if (all(c("Group1", "Group3") %in% input$group)) {
df <- subset(df, (df$Group == "Group1" | df$Group == "Group3"))
return(df)
# if the user only selects Group2 and Group3, it will appear only those columns.
}else if (all(c("Group2", "Group3") %in% input$group)) {
df <- subset(df, (df$Group == "Group2" | df$Group == "Group3"))
return(df)
# if the user only selects Group1
} else if ("Group1" %in% input$group) {
df <- subset(df, (df$Group == "Group1"))
return(df)
# if the user only selects group2
} else if ("Group2" %in% input$group) {
df <- subset(df, (df$Group == "Group2"))
return(df)
# if the user only selects group3
} else if ("Group3" %in% input$group) {
df <- subset(df, (df$Group == "Group3"))
return(df)
# if the user doesn't select anything.
} else {
return(df)
}
}else{
df$Group <- NULL
return(df)
}
})
mydata <- reactive({
req(input$submit)
if(input$log2 == TRUE){
data <- data()
cols <- sapply(data, is.numeric)
data[cols] <- lapply(data[cols], function(x) log2(x 1))
}
else{
data <- data()
}
return(data)
})
draw_bp <- reactive({
if(ncol(mydata())==2){
bp <- ggplot(mydata(), aes(x=groups_age, y=Gene))
geom_boxplot(aes(fill=groups_age), alpha = input$alpha)
labs(fill = "groups_age")
if((input$Kruskalpval == "TRUE") && (input$changeKW==FALSE)){
pval <- mydata() %>%
summarize(Kruskal_pvalue = kruskal.test(Gene ~ groups_age)$p.value)
bp <- bp geom_text(data=pval, aes(x=2, y=max(mydata()$Gene)-10, label=paste0("Kruskal-Wallis\n p = ",Kruskal_pvalue)))
}
if((input$Kruskalpval == "TRUE") && (input$changeKW==TRUE)){
pval <- mydata() %>%
summarize(Kruskal_pvalue = kruskal.test(Gene ~ groups_age)$p.value)
bp <- bp geom_text(data=pval, aes(x=input$X_axis, y=input$Y_axis, label=paste0("Kruskal-Wallis\n p = ",Kruskal_pvalue)))
}
return(bp)
}
else{
bp <- ggplot(mydata(), aes(x=groups_age, y=Gene))
geom_boxplot(aes(fill=groups_age), alpha=input$alpha)
facet_grid(. ~ Group)
if((input$Kruskalpval == "TRUE") && (input$changeKW==FALSE)){
pval <- mydata() %>%
group_by(Group) %>%
summarize(Kruskal_pvalue = kruskal.test(Gene ~ groups_age)$p.value)
bp <- bp geom_text(data=pval, aes(x=2, y=max(mydata()$Gene)-10, label=paste0("Kruskal-Wallis\n p = ",Kruskal_pvalue)))
}
if((input$Kruskalpval == "TRUE") && (input$changeKW==TRUE)){
pval <- mydata() %>%
group_by(Group) %>%
summarize(Kruskal_pvalue = kruskal.test(Gene ~ groups_age)$p.value)
bp <- bp geom_text(data=pval, aes(x=input$X_axis, y=input$Y_axis, label=paste0("Kruskal-Wallis\n p = ",Kruskal_pvalue)))
}
return(bp)
}
})
v <- reactiveValues()
observeEvent(input$show_plot | input$show_plot_2, {
v$plot <- draw_bp()
})
output$boxplots <- renderPlot({
req(input$submit)
if (is.null(v$plot)) return()
v$plot
})
}
shinyApp(ui = ui, server = server)
CodePudding user response:
As @YBS suggested in the comments, we can use eventReactive when creating the reactive data:
data <- eventReactive(c(input$show_plot, input$show_plot_2), {.....})
Complete code:
library(shiny)
library(dplyr)
library(ggplot2)
ui <- fluidPage(
titlePanel("My app"),
sidebarLayout(
sidebarPanel(
tabsetPanel(
tabPanel(
"Tab1",
checkboxInput("log2", "Log2 transformation", value = FALSE),
actionButton("submit", "Submit")
),
tabPanel(
"Tab2",
radioButtons(
inputId = "plot_type", label = "I want to see the plot of:",
c(
"All the samples" = "all_samples",
"Groups" = "samples_group"
)
),
conditionalPanel(
condition = "input.plot_type == 'samples_group'",
style = "margin-left: 20px;",
checkboxGroupInput("group", "Choose the group:",
choices = c("Group1", "Group2", "Group3")
)
),
actionButton("show_plot", "See the plot")
),
tabPanel(
"Tab3",
numericInput("alpha", "Opacity of the plot", value = 0.2),
checkboxInput(inputId = "Kruskalpval", label = "Show the Kruskal Wallis p-value", value = FALSE),
conditionalPanel(
condition = "input.Kruskalpval == '1'",
style = "margin-left: 20px;",
checkboxInput(inputId = "changeKW", "I want to change the place of the value", value = FALSE),
conditionalPanel(
condition = "input.changeKW == '1'",
numericInput(inputId = "X_axis", "X_axis:", value = 2),
numericInput(inputId = "Y_axis", "Y_axis:", value = 70)
)
),
actionButton("show_plot_2", "See the plot")
)
)
),
mainPanel(
plotOutput("boxplots")
)
)
)
server <- function(input, output) {
set.seed(1234)
Gene <- floor(runif(25, min=0, max=101))
groups_age <- floor(runif(25, min=18, max=75))
Group <- c("Group1", "Group1", "Group3", "Group2", "Group1", "Group3", "Group2", "Group2", "Group2", "Group1", "Group1", "Group3", "Group1", "Group2", "Group1", "Group2", "Group3", "Group1", "Group3", "Group3", "Group2", "Group1", "Group3", "Group3","Group2")
data <- eventReactive(c(input$show_plot, input$show_plot_2), {
df <- data.frame(Gene, Group, groups_age)
mybreaks <- seq(min(df$groups_age)-1, to=max(df$groups_age) 10, by=10)
df$groups_age <- cut(df$groups_age, breaks = mybreaks, by=10)
if(input$plot_type == "samples_group"){
# if the user selects everything, it will take everything.
if(all(c("Group1", "Group2", "Group3") %in% input$group)){
return(df)
# if the user only selects group1 and group2, it will appear only those columns.
}else if (all(c("Group1", "Group2") %in% input$group)) {
df <- subset(df, (df$Group == "Group1" | df$Group == "Group2"))
return(df)
# if the user only selects group1 and group3, it will appear only those columns.
}else if (all(c("Group1", "Group3") %in% input$group)) {
df <- subset(df, (df$Group == "Group1" | df$Group == "Group3"))
return(df)
# if the user only selects Group2 and Group3, it will appear only those columns.
}else if (all(c("Group2", "Group3") %in% input$group)) {
df <- subset(df, (df$Group == "Group2" | df$Group == "Group3"))
return(df)
# if the user only selects Group1
} else if ("Group1" %in% input$group) {
df <- subset(df, (df$Group == "Group1"))
return(df)
# if the user only selects group2
} else if ("Group2" %in% input$group) {
df <- subset(df, (df$Group == "Group2"))
return(df)
# if the user only selects group3
} else if ("Group3" %in% input$group) {
df <- subset(df, (df$Group == "Group3"))
return(df)
# if the user doesn't select anything.
} else {
return(df)
}
}else{
df$Group <- NULL
return(df)
}
})
mydata <- reactive({
req(input$submit)
if(input$log2 == TRUE){
data <- data()
cols <- sapply(data, is.numeric)
data[cols] <- lapply(data[cols], function(x) log2(x 1))
}
else{
data <- data()
}
return(data)
})
draw_bp <- reactive({
if(ncol(mydata())==2){
bp <- ggplot(mydata(), aes(x=groups_age, y=Gene))
geom_boxplot(aes(fill=groups_age), alpha = input$alpha)
labs(fill = "groups_age")
if((input$Kruskalpval == "TRUE") && (input$changeKW==FALSE)){
pval <- mydata() %>%
summarize(Kruskal_pvalue = kruskal.test(Gene ~ groups_age)$p.value)
bp <- bp geom_text(data=pval, aes(x=2, y=max(mydata()$Gene)-10, label=paste0("Kruskal-Wallis\n p = ",Kruskal_pvalue)))
}
if((input$Kruskalpval == "TRUE") && (input$changeKW==TRUE)){
pval <- mydata() %>%
summarize(Kruskal_pvalue = kruskal.test(Gene ~ groups_age)$p.value)
bp <- bp geom_text(data=pval, aes(x=input$X_axis, y=input$Y_axis, label=paste0("Kruskal-Wallis\n p = ",Kruskal_pvalue)))
}
return(bp)
}
else{
bp <- ggplot(mydata(), aes(x=groups_age, y=Gene))
geom_boxplot(aes(fill=groups_age), alpha=input$alpha)
facet_grid(. ~ Group)
if((input$Kruskalpval == "TRUE") && (input$changeKW==FALSE)){
pval <- mydata() %>%
group_by(Group) %>%
summarize(Kruskal_pvalue = kruskal.test(Gene ~ groups_age)$p.value)
bp <- bp geom_text(data=pval, aes(x=2, y=max(mydata()$Gene)-10, label=paste0("Kruskal-Wallis\n p = ",Kruskal_pvalue)))
}
if((input$Kruskalpval == "TRUE") && (input$changeKW==TRUE)){
pval <- mydata() %>%
group_by(Group) %>%
summarize(Kruskal_pvalue = kruskal.test(Gene ~ groups_age)$p.value)
bp <- bp geom_text(data=pval, aes(x=input$X_axis, y=input$Y_axis, label=paste0("Kruskal-Wallis\n p = ",Kruskal_pvalue)))
}
return(bp)
}
})
v <- reactiveValues()
observeEvent(input$show_plot | input$show_plot_2, {
v$plot <- draw_bp()
})
output$boxplots <- renderPlot({
req(input$submit)
if (is.null(v$plot)) return()
v$plot
})
}
shinyApp(ui = ui, server = server)
CodePudding user response:
You need to use isolate() for the numeric inputs so that they do not update the position of KW without clicking on the actionButton. Also, no need of observeEvent(). Try this
draw_bp <- eventReactive(c(input$show_plot, input$show_plot_2), {
if(ncol(mydata())==2){
bp <- ggplot(mydata(), aes(x=groups_age, y=Gene))
geom_boxplot(aes(fill=groups_age), alpha = input$alpha)
labs(fill = "groups_age")
if((input$Kruskalpval == "TRUE") && (input$changeKW==FALSE)){
pval <- mydata() %>%
summarize(Kruskal_pvalue = kruskal.test(Gene ~ groups_age)$p.value)
bp <- bp geom_text(data=pval, aes(x=2, y=max(mydata()$Gene)-10, label=paste0("Kruskal-Wallis\n p = ",Kruskal_pvalue)))
}
if((input$Kruskalpval == "TRUE") && (input$changeKW==TRUE)){
pval <- mydata() %>%
summarize(Kruskal_pvalue = kruskal.test(Gene ~ groups_age)$p.value)
bp <- bp geom_text(data=pval, aes(x=isolate(input$X_axis), y=isolate(input$Y_axis), label=paste0("Kruskal-Wallis\n p = ",Kruskal_pvalue)))
}
return(bp)
}
else{
bp <- ggplot(mydata(), aes(x=groups_age, y=Gene))
geom_boxplot(aes(fill=groups_age), alpha=input$alpha)
facet_grid(. ~ Group)
if((input$Kruskalpval == "TRUE") && (input$changeKW==FALSE)){
pval <- mydata() %>%
group_by(Group) %>%
summarize(Kruskal_pvalue = kruskal.test(Gene ~ groups_age)$p.value)
bp <- bp geom_text(data=pval, aes(x=2, y=max(mydata()$Gene)-10, label=paste0("Kruskal-Wallis\n p = ",Kruskal_pvalue)))
}
if((input$Kruskalpval == "TRUE") && (input$changeKW==TRUE)){
pval <- mydata() %>%
group_by(Group) %>%
summarize(Kruskal_pvalue = kruskal.test(Gene ~ groups_age)$p.value)
bp <- bp geom_text(data=pval, aes(x=isolate(input$X_axis), y=isolate(input$Y_axis), label=paste0("Kruskal-Wallis\n p = ",Kruskal_pvalue)))
}
return(bp)
}
})
v <- reactiveValues()
observeEvent(input$show_plot | input$show_plot_2, {
v$plot <- draw_bp()
})
output$boxplots <- renderPlot({
req(input$submit)
# if (is.null(v$plot)) return()
# v$plot
draw_bp()
})
