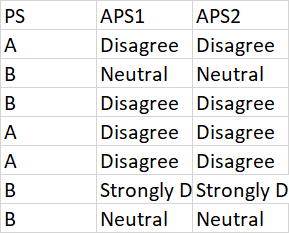
As shown in image, i have a .csv file. I am trying to create charts. I pass dataframe from this image to a function multi_bar(dt)
The code uses a FOR LOOP and take column 1 (PS) and column 2 (APS1) in one go and then using column 1 (PS) and then column 2(APS2) by using for loop and dplyr. Basically I need column 1 in each iteration but the other column will keep changing from 2,3 and so on. The code here doesn't show code for chart but the error is coming when the code reaches column 3 (i=3) in for loop iteration. It works for i=2.
multi_bar=function(dt)
{
library(dplyr)
library(ggplot2)
for(i in 2:ncol(dt))
{
col1 = names(dt)[1]
col2=names(dt[i])
#df=select(x,x[,1], x[,i])
df=dt[,c(1,i)]
df1=df%>%
group_by(df[,1], df[,i])%>%
summarize(n=n())
colnames(df1) <- c(col1,col2,'Count')
col3=names(df1)[3]
ls=list(df1,col1,col2,col3,ncol(dt))
}
return(ls)
}
when I pass dataframe to this function multi_bar, as shown in image (after reading csv file), the following error occurs
Error: Problem adding computed columns in group_by().
x Problem with mutate() input ..2.
i ..2 = df[, i].
x undefined columns selected
Run rlang::last_error() to see where the error occurred.
In addition: Warning message:
Error: Problem adding computed columns in group_by().
x Problem with mutate() input ..2.
i ..2 = df[, i].
x undefined columns selected
Run rlang::last_error() to see where the error occurred.
CodePudding user response:
I think you'll greatly benefit by bringing the data in long (tidy) format. Then you can use count to count number of times each value occurs in the column.
We don't have your data to test but it would be something like this -
library(dplyr)
library(tidyr)
df <- dt %>%
pivot_longer(cols = -PS) %>%
count(PS, name, value, name = 'Count')
CodePudding user response:
While I agree with the suggestion above of Ronak Shah, it maybe doesn't answer your question, as to how to fix the error you are having.
To access directly the index positions you need you can use the following snippet in your group_by statement:
df1 <- df %>%
group_by(.[[1]], .[[2]]) %>%
summarize(n=n())
If you want to cut down on a step and rely on the looping variable you can do
df1 <- dt %>%
group_by(.[[1]], .[[i]]) %>%
summarize(n=n())
If you have the column names, like you have created already you can use the function !! before your column names in the group_by statement like so:
df1=df %>%
group_by(!!col1, !!col2)%>%
summarize(n=n())
I can imagine that you used your code, outside of the function or outside of the loop and the code worked fine, when you put it into a loop, or indeed into the function, you change the scope of the variables. Coupled with the fact that dplyr uses tidy evaluation, this can get messy.
!! is used to force early evaluation, which is what you would need here so that the variables contained values are used rather than the variable names themselves.