library(ggplot2)
devtools::install_github("google/CausalImpact")
library(CausalImpact)
## note that I took this example code from the package documentation up until I customize the plot
#create data
set.seed(1)
x1 <- 100 arima.sim(model = list(ar = 0.999), n = 100)
y <- 1.2 * x1 rnorm(100)
y[71:100] <- y[71:100] 10
data <- cbind(y, x1)
#causal impact analysis
> pre.period <- c(1, 70)
> post.period <- c(71, 100)
> impact <- CausalImpact(data, pre.period, post.period)
#graph
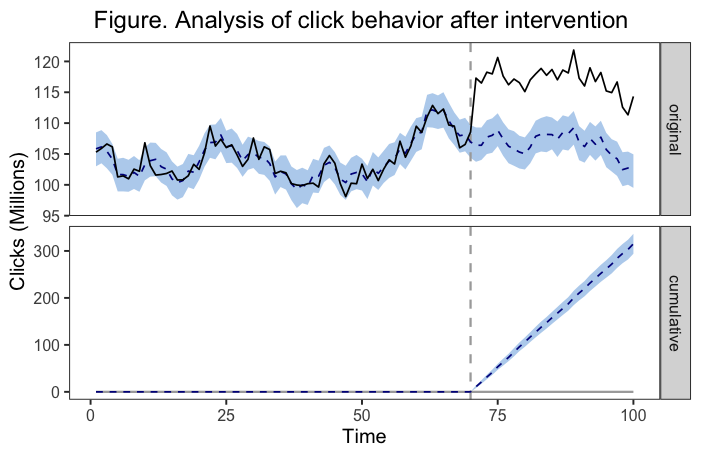
example<-plot(impact, c("original", "cumulative"))
labs(
x = "Time",
y = "Clicks (Millions)",
title = "Figure. Analysis of click behavior after intervention.")
theme(plot.title = element_text(hjust = 0.5),
plot.caption = element_text(hjust = 0),
panel.background = element_rect(fill = "transparent"), # panel bg
plot.background = element_rect(fill = "transparent", color = NA), # plot bg
panel.grid.major = element_blank(), # get rid of major grid
panel.grid.minor = element_blank()) # get rid of minor grid
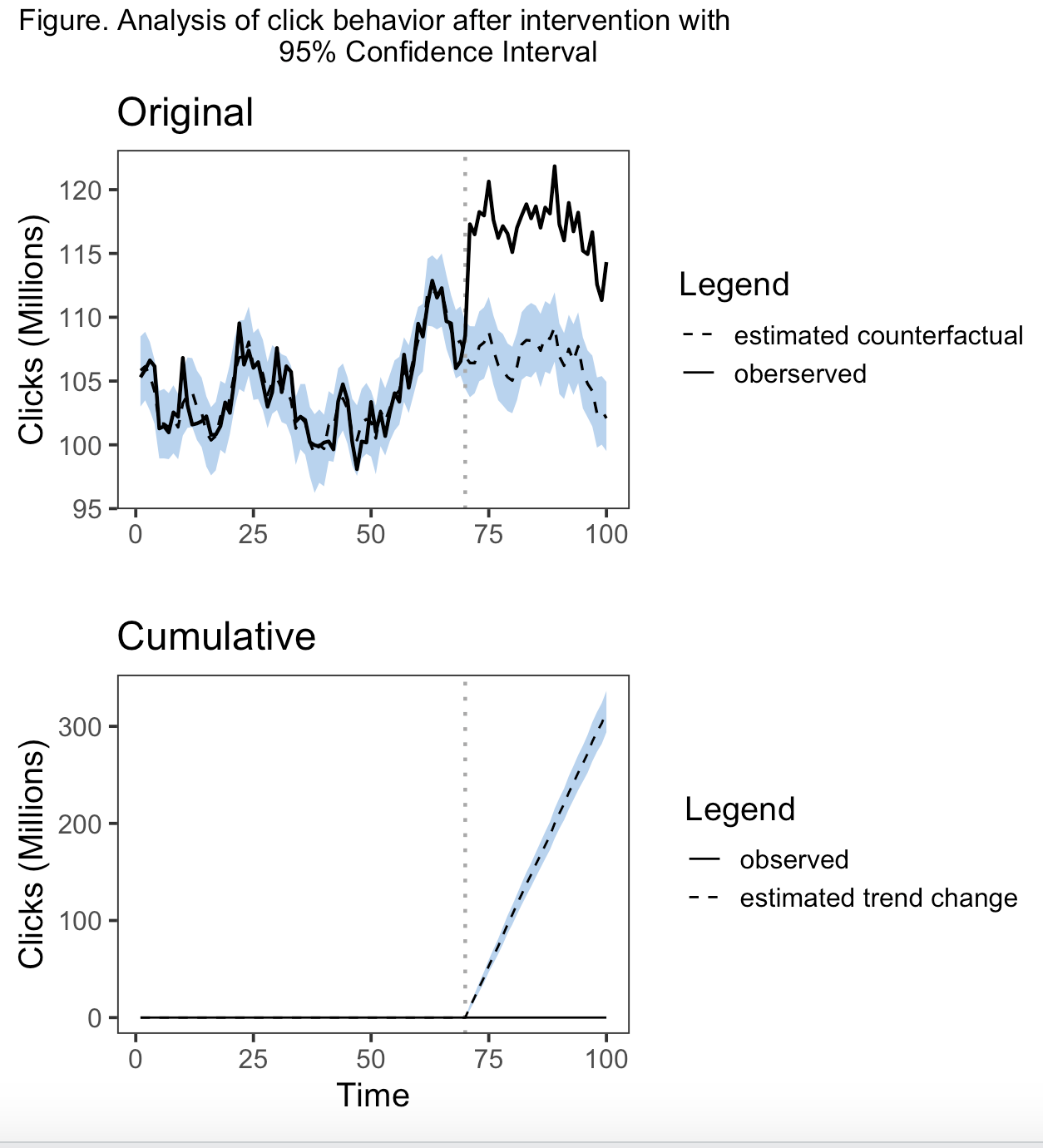
In my head, the solution I'd like is to have a legend for each panel of the plot. The first legend (next to the 'original' panel) would show a solid line represents the observed data, the dotted line represents the estimated counterfactual, and the colored band represents the 95% CrI around the estimated counterfactual. The second legend (next to the 'cumulative' panel) would show the dotted line represents the estimated change in trend associated with the intervention and the colored band again represents the 95% CrI around the estimation. Maybe there's a better solution than that, but that's what I've thought of.
CodePudding user response:
With a bit more work, here is another version that is closer to the original aesthetic. Instructions as per above with the addition of library(grid). Code could use some clean up, but I just focused on getting a graphic as close to the one you specified. I brought some of the themes parameters back outside the function.
CreateImpactPlot <- function(impact, metrics = c("original", "pointwise",
"cumulative")) {
# Creates a plot of observed data and counterfactual predictions.
#
# Args:
# impact: \code{CausalImpact} results object returned by
# \code{CausalImpact()}.
# metrics: Which metrics to include in the plot. Can be any combination of
# "original", "pointwise", and "cumulative".
#
# Returns:
# A ggplot2 object that can be plotted using plot().
# Create data frame of: time, response, mean, lower, upper, metric
data <- CreateDataFrameForPlot(impact)
# Select metrics to display (and their order)
assert_that(is.vector(metrics))
metrics <- match.arg(metrics, several.ok = TRUE)
data <- data[data$metric %in% metrics, , drop = FALSE]
data$metric <- factor(data$metric, metrics)
# Initialize plot
#q <- ggplot(data, aes(x = time)) theme_bw(base_size = 15)
#q <- q xlab("") ylab("")
#if (length(metrics) > 1) {
# q <- q facet_grid(metric ~ ., scales = "free_y")
#}
q1 <- ggplot(data, aes(x = time)) theme_bw(base_size = 15)
q1 <- q1 xlab("") ylab("")
q2 <- ggplot(data, aes(x = time)) theme_bw(base_size = 15)
q2 <- q2 xlab("") ylab("")
q3 <- ggplot(data %>%
filter(metric == "cumulative") %>%
mutate(metric = factor(metric, levels = c("cumulative"))), aes(x = time)) theme_bw(base_size = 15)
q3 <- q3 xlab("") ylab("")
# Add prediction intervals
#q <- q geom_ribbon(aes(ymin = lower, ymax = upper),
# data, fill = "slategray2")
q1 <- q1 geom_ribbon(data = data %>%
filter(metric == "original") %>%
mutate(metric = factor(metric, levels = c("original"))), aes(x = time, ymin = lower, ymax = upper),
fill = "slategray2")
q2 <- q2 geom_ribbon(data = data %>%
filter(metric == "pointwise") %>%
mutate(metric = factor(metric, levels = c("pointwise"))), aes(x = time, ymin = lower, ymax = upper),
fill = "slategray2")
q3 <- q3 geom_ribbon(data = data %>%
filter(metric == "cumulative") %>%
mutate(metric = factor(metric, levels = c("cumulative"))), aes(x = time, ymin = lower, ymax = upper),
fill = "slategray2")
# Add pre-period markers
xintercept <- CreatePeriodMarkers(impact$model$pre.period,
impact$model$post.period,
time(impact$series))
#q <- q geom_vline(xintercept = xintercept,
# colour = "darkgrey", size = 0.8, linetype = "dashed")
q1 <- q1 geom_vline(xintercept = xintercept,
colour = "darkgrey", size = 0.8, linetype = "dotted")
q2 <- q2 geom_vline(xintercept = xintercept,
colour = "darkgrey", size = 0.8, linetype = "dotted")
q3 <- q3 geom_vline(xintercept = xintercept,
colour = "darkgrey", size = 0.8, linetype = "dotted")
data_long <- data %>%
tidyr::pivot_longer(cols = c("baseline", "mean", "response"), names_to = "variable",
values_to = "value", values_drop_na = TRUE)
# Add zero line to pointwise and cumulative plot
#q <- q geom_line(aes(y = baseline),
# colour = "darkgrey", size = 0.8, linetype = "solid",
# na.rm = TRUE)
q1 <- q1 geom_line(data = data_long %>% dplyr::filter(metric == "original"),
aes(x = time, y = value, linetype = variable, group = variable,
size = variable),
na.rm = TRUE)
scale_linetype_manual(guide = "Legend", labels = c("estimated counterfactual", "oberserved"),
values = c("dashed", "solid"))
scale_size_manual(values = c(0.6, 0.8))
scale_color_manual(values = c("darkblue", "darkgrey"))
theme(legend.position = "right", axis.text.x = element_blank(), axis.title.y = element_blank())
guides(linetype = guide_legend("Legend", nrow=2), size = "none", color = "none")
facet_wrap(~metric[1], strip.position = "right", drop = TRUE)
#q2 <- q2 geom_line(data = data_long %>% dplyr::filter(metric == "pointwise"),
# aes(x = time, y = value, linetype = Line, group = Line),
# na.rm = TRUE)
# scale_linetype_manual(title = "Legend", labels = c("estimated counterfactual", "observed"),
# values = c("dashed", "solid"))
# scale_size_manual(values = c(0.6, 0.8))
# scale_color_manual(values = c("darkblue", "darkgrey"))
# theme(legend.position = "right")
# guides(linetype = guide_legend("Legend", nrow=2), size = "none", color = "none")
# labs(title = "Pointwise", y = "Clicks (Millions)")
q3 <- q3 geom_line(d = data_long %>%
filter(metric == "cumulative") %>%
mutate(metric = factor(metric, levels = c("cumulative"))),
aes(x = time, y = value, linetype = variable, group = variable),
na.rm = TRUE)
scale_linetype_manual(labels = c("observed", "estimated trend change"),
values = c("solid", "dashed"))
theme(legend.position = "right", axis.title.y = element_blank())
guides(linetype = guide_legend("Legend", nrow=2))
labs(x = "Time")
facet_wrap(~metric, strip.position = "right", drop = TRUE)
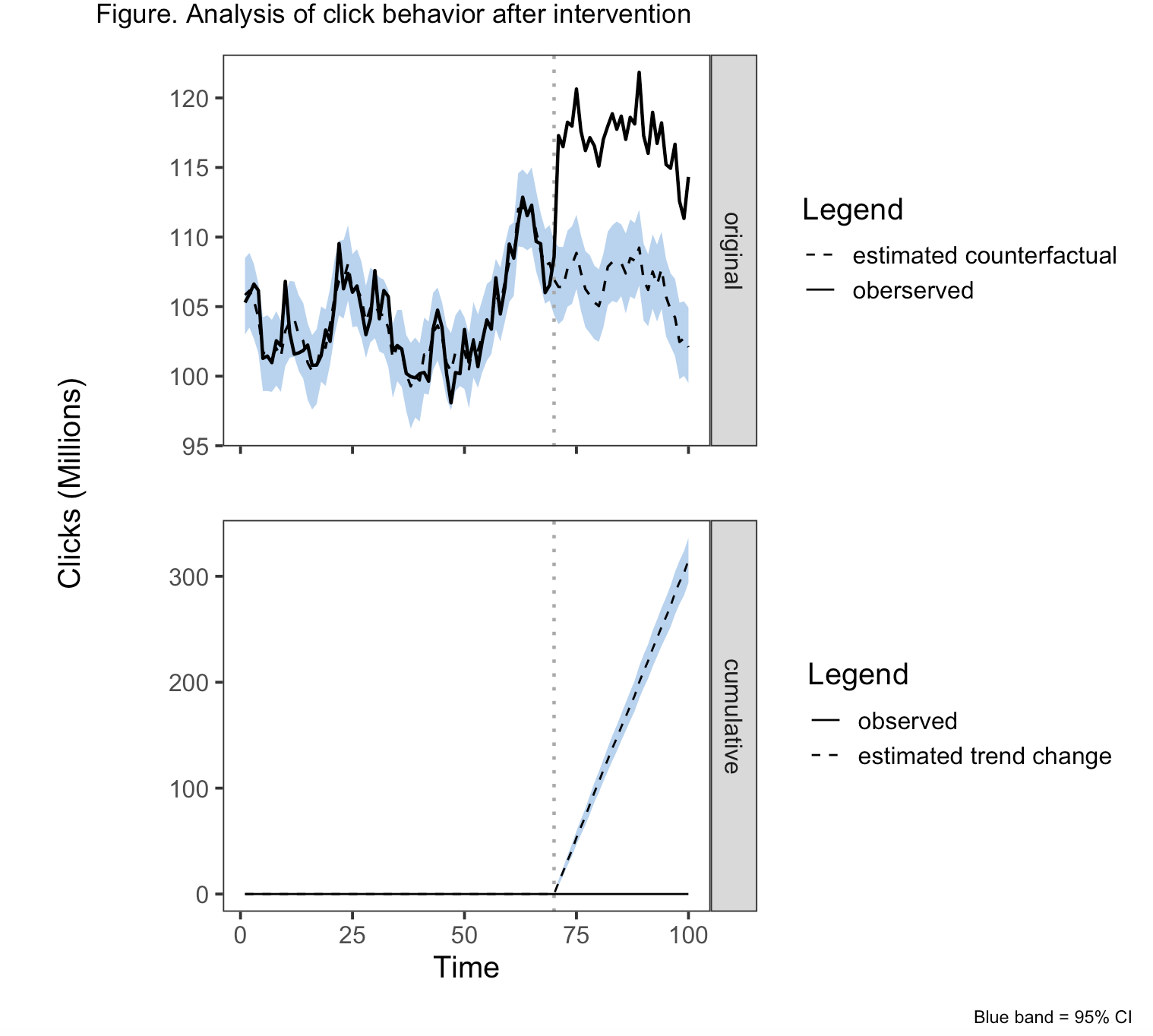
g1 <- grid::textGrob("Clicks (Millions)", rot = 90, gp=gpar(fontsize = 15), x= 0.85)
wrap_elements(g1) | (q1/q3)
patchwork <- wrap_elements(g1) | (q1/q3)
q <- patchwork
# Add point predictions
#q <- q geom_line(aes(y = mean), data,
# size = 0.6, colour = "darkblue", linetype = "dashed",
# na.rm = TRUE)
# Add observed data
#q <- q geom_line(aes(y = response), size = 0.6, na.rm = TRUE)
return(q)
}
## Use this to run the plot ##
plot(impact, c("original", "cumulative"))
plot_annotation(title = "Figure. Analysis of click behavior after intervention"
, theme = theme(plot.title = element_text(hjust = 0.5)),
caption = "Blue band = 95% CI") &
theme(
panel.background = element_rect(fill = "transparent"), # panel bg
plot.background = element_rect(fill = "transparent", color = NA), # plot bg
panel.grid.major = element_blank(), # get rid of major grid
panel.grid.minor = element_blank())