I have a dataframe called cells, with cells in rows and samples in columns, here is a dput sample:
structure(c(8.10937548981953e-20, 0.095381661829093, 0.054868371418562,
0.0523687378840825, 0.0100173293159538, 0.0332395245437795, 3.37811149975583e-20,
0.048191378909587, 0.13314908462763, 0, 0.00612878313809124,
0, 0.00209460699328093, 0.205599458004829, 0.318048653115709,
4.21796249339787e-05, 0.00844407692255898, 0, 0.00613007026042523,
0.0300024082993193, 0.0405191646567986, 0.00654087887823056,
0.0111094954094255, 1.30617589099212e-19, 0.0398730537850546,
0.0390946117756341, 0.239413780024853, 2.07521807718399e-19,
0.00116980239850497, 0, 0, 0.00971921247886335, 0.0588607291731613,
3.8563512241696e-21, 0.00247621905821516, 0), .Dim = c(6L, 6L
), .Dimnames = list(c("Adipocytes", "B-cells", "Basophils", "CD4 memory T-cells",
"CD4 naive T-cells", "CD4 T-cells"), c("Pt1", "Pt10", "Pt103",
"Pt106", "Pt11", "Pt17")))
number of rows in cells is 38 and number of columns is 49.
My target cell type that I want to build a boxplot for is: CD4 memory T-cells
In addition, have another data frame called Metadata, which contains the response column, if a sample responded to a certain therapy or not - Metadata$Benefit. So for each sample in cells I can know whether they responded or not.
Goal: make a boxplot with Response/No Response in the x axis and values on the y axis, and I need the boxplot with the indiviudal data points on it.
My code:
cells %>%
ggplot( aes(x = as.factor(Metadata$Benefit), y = as.numeric(cells['CD4 memory T-cells',]), fill= c('red','lightblue')))
geom_boxplot()
scale_fill_viridis(discrete = TRUE, alpha=0.6)
geom_jitter(color="black", size=0.4, alpha=0.9)
theme_ipsum()
theme(
legend.position="none",
plot.title = element_text(size=11)
)
ggtitle("A boxplot with jitter")
xlab("")
Error: Aesthetics must be either length 1 or the same as the data (38): x, y and fill
I don't get what's the problem because both length(as.factor(Metadata$Benefit)) and length(as.numeric(cells['CD4 memory T-cells',])) is 49, it's the same so what's the problem?
CodePudding user response:
There are a few issues with your attempt here as mentioned in the comments. You can't take x and y aesthetics from different data sources so you should merge them first. This also means rotating (for which I used t() while it was still a matrix) the original data prior to merging so each sample is a row. To demonstrate this I made up some response metadata.
library(tidyverse)
d <- structure(c(8.10937548981953e-20, 0.095381661829093, 0.054868371418562, 0.0523687378840825, 0.0100173293159538, 0.0332395245437795, 3.37811149975583e-20, 0.048191378909587, 0.13314908462763, 0, 0.00612878313809124, 0, 0.00209460699328093, 0.205599458004829, 0.318048653115709, 4.21796249339787e-05, 0.00844407692255898, 0, 0.00613007026042523, 0.0300024082993193, 0.0405191646567986, 0.00654087887823056, 0.0111094954094255, 1.30617589099212e-19, 0.0398730537850546, 0.0390946117756341, 0.239413780024853, 2.07521807718399e-19, 0.00116980239850497, 0, 0, 0.00971921247886335, 0.0588607291731613, 3.8563512241696e-21, 0.00247621905821516, 0), .Dim = c(6L, 6L), .Dimnames = list(c("Adipocytes", "B-cells", "Basophils", "CD4 memory T-cells", "CD4 naive T-cells", "CD4 T-cells"), c("Pt1", "Pt10", "Pt103", "Pt106", "Pt11", "Pt17")))
metadata <- data.frame(sample = c("Pt1", "Pt10", "Pt103", "Pt106", "Pt11", "Pt17"), response = c(T, T, T, F, F, F))
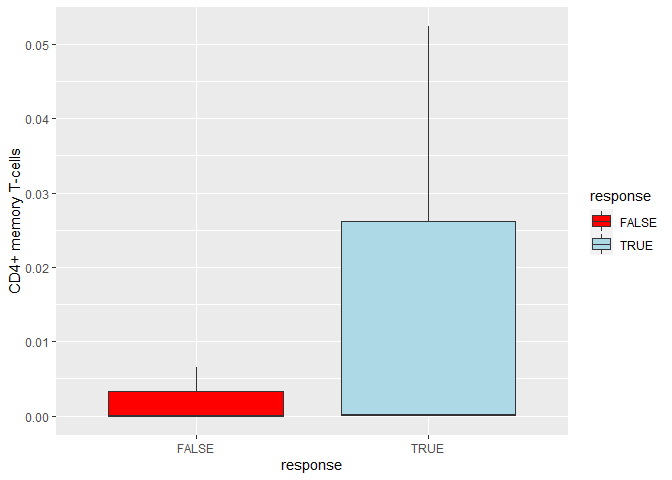
d %>%
t() %>%
as.data.frame() %>%
rownames_to_column("sample") %>%
right_join(metadata, .) %>%
ggplot(aes(x = response, y = `CD4 memory T-cells`))
geom_boxplot(aes(fill = response))
scale_fill_manual(values = c('red','lightblue'))
#> Joining, by = "sample"
Created on 2022-02-16 by the reprex package (v2.0.1)
