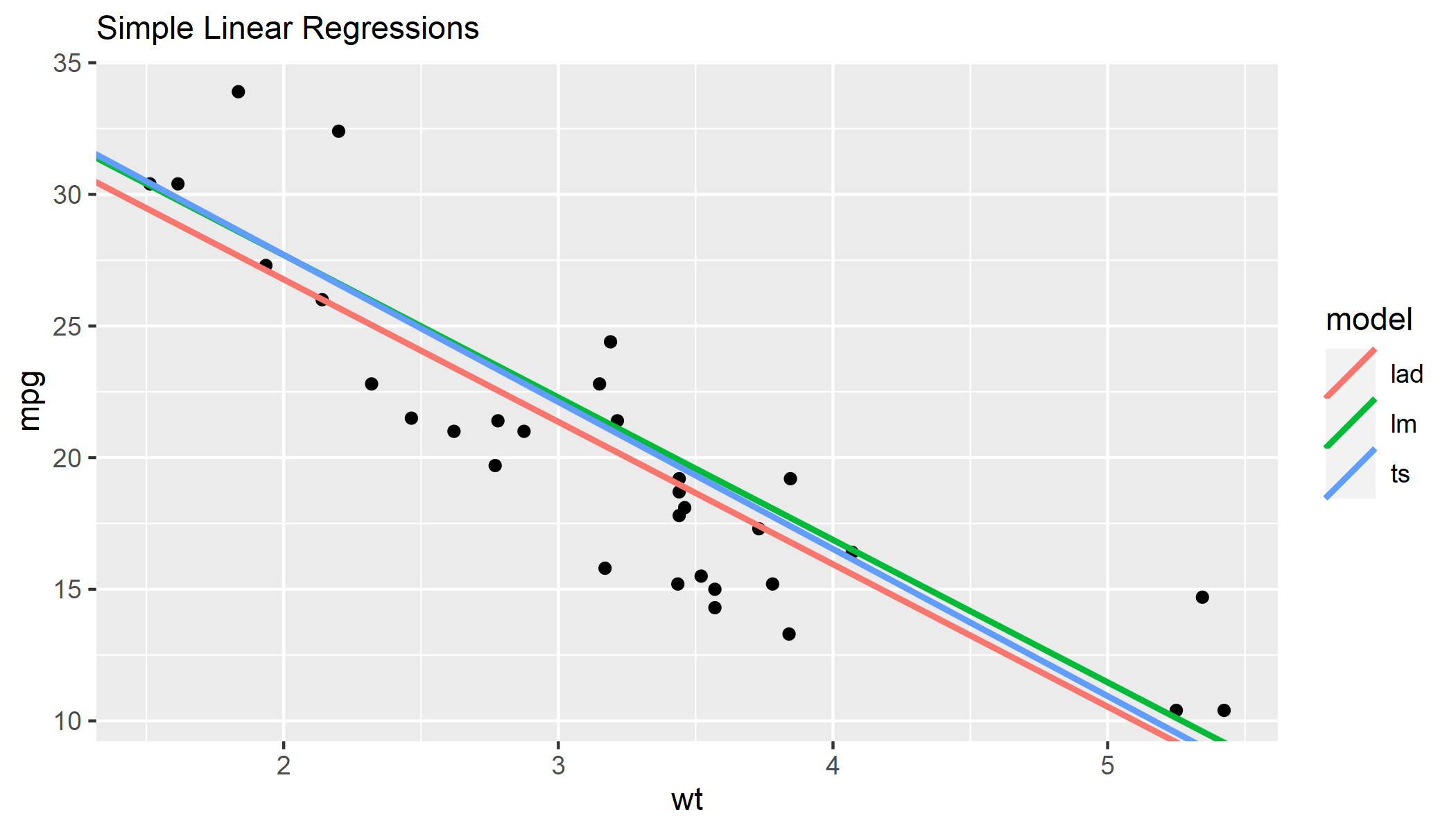
I'm trying to plot different simple linear regression estimates on the same coordinate plane to understand something of the differences between different methods. But my question is about adding these lines in R code and not about the statistics of the differences lines.
Here I'm using the mtcars dataset. And I'm using the mblm and quantreq packages to come up with different regression equations or, more specifically, the parameters for the slope and intercept for different simple linear regression estimates.
The OLS estimate I add using the geom_smooth() function and specifying the method argument. I could add the slope and intercept using geom_abline() after creating a linear model object; that's another option.
The Theil-Sen and median least squares deviation estimates I'm creating a model first for each using the respective packages. Then I'm adding the slope and intercept using geom_abline().
So now I've added the lines manually. But how can I create a key or legend in ggplot to show these different lines? ggplot() adds the key automatically when geom_smooth() is separated into different groups. But I don't think it adds a legend for geom_abline. And anyway my plot uses a mixture of both. Any ideas? I've never had to add more own key in this way.
library(mblm)
ts_fit <- mblm(mpg ~ wt, data = mtcars)
library(quantreg)
lad_fit <- rq(mpg ~ wt, data = mtcars)
ggplot(mtcars, aes(x = wt, y = mpg))
labs(subtitle = "Simple Linear Regressions")
geom_point()
geom_smooth(method = 'lm', se = FALSE, color = '#376795')
geom_abline(intercept = coef(ts_fit)[1], slope = coef(ts_fit)[2], color = '#f7aa58', size = 1)
geom_abline(intercept = coef(lad_fit)[1], slope = coef(lad_fit)[2], color = '#72bcd5', size = 1)
CodePudding user response:
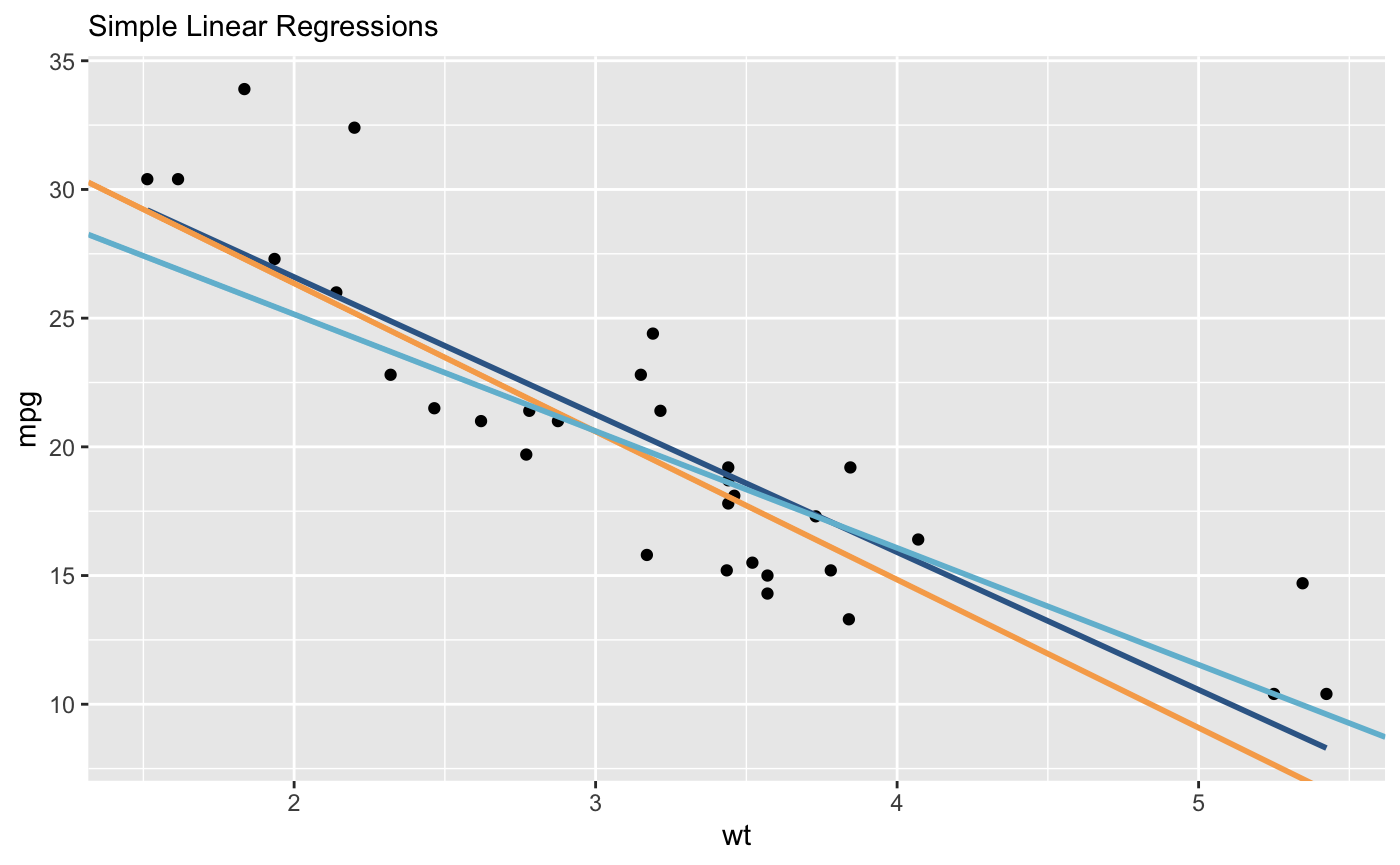
Rather than adding a separate geom for each model, I would create a dataframe including the intercept and slope for all models. Then you can pass this to a single geom_abline() and map color to the different models.
Note, I don't have {mblm} or {quantreg} installed, so I ran lm() on different subsets of mtcars as an approximation.
library(tidyverse)
# create dataframe with model coefficients
models <- data.frame(
lm = coef(lm(mpg ~ wt, data = mtcars[1:20,])),
ts = coef(lm(mpg ~ wt, data = mtcars[7:26,])),
lad = coef(lm(mpg ~ wt, data = mtcars[11:32,]))
) %>%
t() %>%
as_tibble(rownames = "model") %>%
rename_with(~ c("model", "intercept", "slope"))
models
# # A tibble: 3 x 3
# model intercept slope
# <chr> <dbl> <dbl>
# 1 lm 38.5 -5.41
# 2 ts 38.9 -5.59
# 3 lad 37.6 -5.41
# specify ggplot, passing `mtcars` to `geom_point()` and `models` to `geom_abline()`
ggplot()
labs(subtitle = "Simple Linear Regressions")
geom_point(data = mtcars, aes(wt, mpg))
geom_abline(
data = models,
aes(intercept = intercept, slope = slope, color = model),
size = 1
)