I found some other similarly titled questions but didn't find the answer.
My text is:
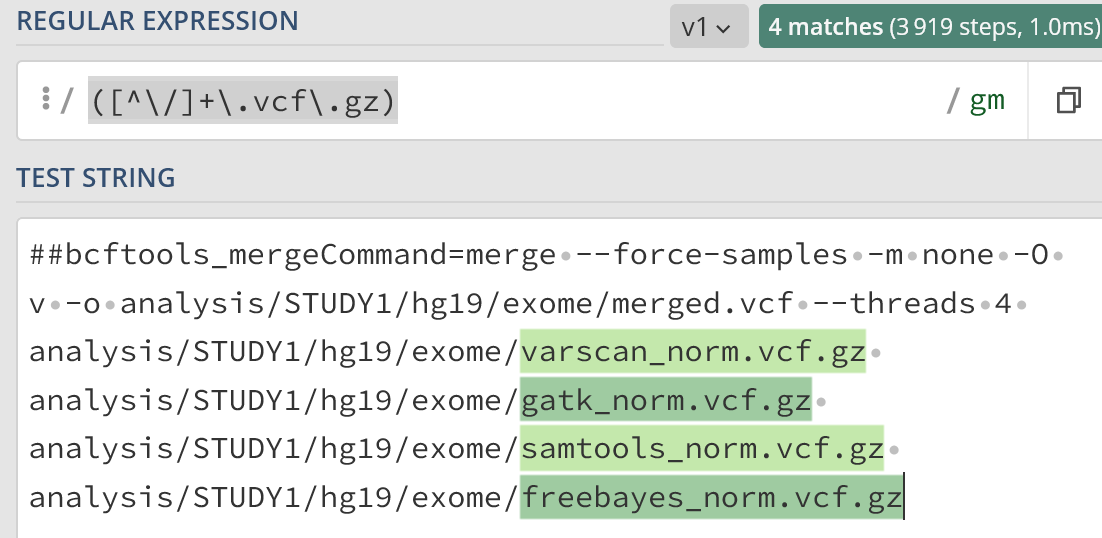
##bcftools_mergeCommand=merge --force-samples -m none -O v -o analysis/STUDY1/hg19/exome/merged.vcf --threads 4 analysis/STUDY1/hg19/exome/varscan_norm.vcf.gz analysis/STUDY1/hg19/exome/gatk_norm.vcf.gz analysis/STUDY1/hg19/exome/samtools_norm.vcf.gz analysis/STUDY1/hg19/exome/freebayes_norm.vcf.gz
I want the names of the .vcf.gz files.
Sed gives me:
echo "##bcftools_mergeCommand=merge --force-samples -m none -O v -o analysis/STUDY1/hg19/exome/merged.vcf --threads 4 analysis/STUDY1/hg19/exome/varscan_norm.vcf.gz analysis/STUDY1/hg19/exome/gatk_norm.vcf.gz analysis/STUDY1/hg19/exome/samtools_norm.vcf.gz analysis/STUDY1/hg19/exome/freebayes_norm.vcf.gz" | sed -En 's/\/([^\/] \.vcf\.gz)/\1/g'
with blank results.
Regex101 gives:
https://regex101.com/r/h3OGvN/1
CodePudding user response:
Why not using grep ?
$ data='##bcftools_mergeCommand=merge --force-samples -m none -O v -o analysis/STUDY1/hg19/exome/merged.vcf --threads 4 analysis/STUDY1/hg19/exome/varscan_norm.vcf.gz analysis/STUDY1/hg19/exome/gatk_norm.vcf.gz analysis/STUDY1/hg19/exome/samtools_norm.vcf.gz analysis/STUDY1/hg19/exome/freebayes_norm.vcf.gz'
$ echo $data | grep -Eo [^\/] \.vcf\.gz
varscan_norm.vcf.gz
gatk_norm.vcf.gz
samtools_norm.vcf.gz
freebayes_norm.vcf.gz
-E: Interpret patterns as extended regular expressions.-o: Print only the matched (non-empty) parts.
CodePudding user response:
The regex dialect supported by Regex101 is different from the one sed understands.
Concretely, (take out the superflous g flag and) add a p flag to print the matching lines to fix this specific script; but in the general case, don't rely on a tool which doesn't directly support the one you actually want to use.