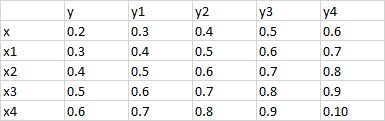
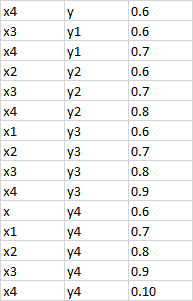
I have a correlation dataframe with 381717 rows and 450 columns and no NA values, and I want to subset this dataframe for all correlations with abs value > 0.6. I have tried multiple things to use lapply and sapply on all rows and columns to subset my dataframe but I end up getting NAs, but I do see that there are a few values which should satisfy this condition.If I could get any leads on how to do this, I would be really grateful.
I know this seems like an easy issue but I am somehow unable to get the right subsetting done and would like your help!
Thanks in advance!
Best regards
Expected output :
CodePudding user response:
x1 = seq(1:7)
x2 = c(2,4,8,5,1,2,3)
y1 = c(9,6,5,4,8,6,4)
y2 = c(1,7,4,5,1,2,2)
df = data.frame(x1,x2,y1,y2)
corr_df = data.frame(cor(df))
corr_df$var = row.names(corr_df)
corr_df1 = reshape2::melt(corr_df, value_name = "Corr")
corr_df1[corr_df1$value > 0.6,]
I have created a dummy dataset and done the subset of correlation dataframe. It might work for you.
CodePudding user response:
Considering a dataframe of correlation values:
corr.vals<-data.frame(x1=runif(5,0,1),
x2=runif(5,0,1),
x3=runif(5,0,1),
x4=runif(5,0,1),
x5=runif(5,0,1))
row.names(corr.vals)<-c("y1","y2","y3","y4","y5")
You should be able to select the values > 0.6, while keeping row and column names, using complete.cases() in a subsetting:
values_06<-corr.vals[complete.cases(corr.vals)>0.6]