I have a problem where I want to render a markdown file via my Shiny app. Iam running RStudio server on Linux Red Hat. I have previously used the code below with success, however it does not work now for some reason.
The issue is that R code is not rendered, see below. When rendering directly via the .Rmd file it works as expected. Both app.R and prot.Rmd are located in the same folder.
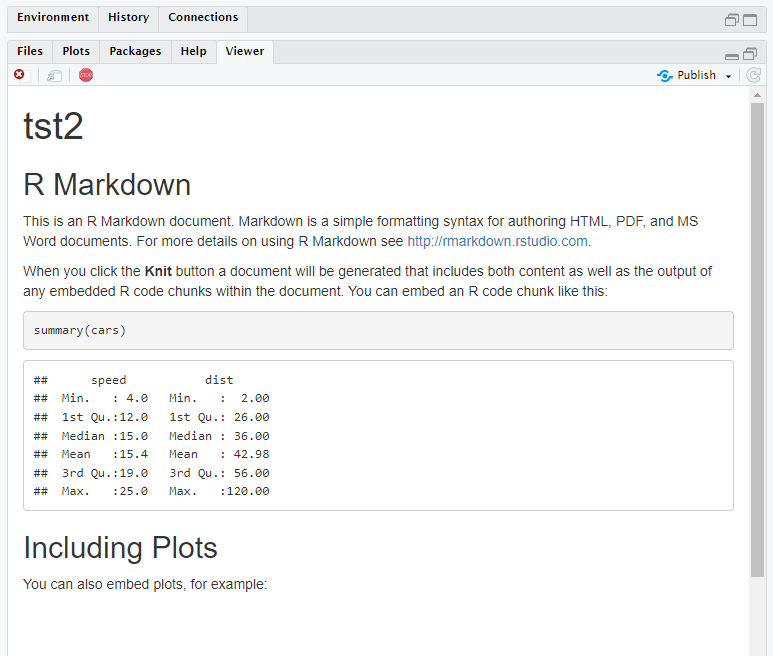
Markdown rendered via Shiny app
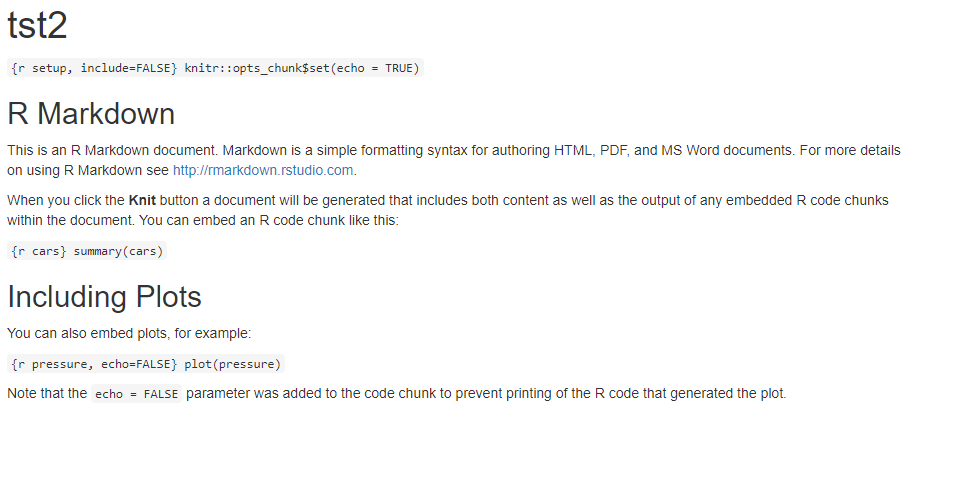 Markdown rendered in RStudio
Markdown rendered in RStudio
Shiny app
library(shiny)
library(rmarkdown)
# Define UI for application that draws a histogram
ui <- fluidPage(
# Application title
titlePanel("Knit"),
downloadButton("prot", "Render file")
)
# Define server logic required to draw a histogram
server <- function(input, output) {
output$prot <- downloadHandler(
# For PDF output, change this to "report.pdf"
filename = "prot.html",
content = function(file) {
report_path <- tempfile(fileext = ".html")
file.copy("prot.Rmd", report_path, overwrite = T)
rmarkdown::render(report_path,
output_file = file,
envir = new.env(parent = globalenv())
)
}
)
}
# Run the application
shinyApp(ui = ui, server = server)
Rmd-file
---
title: "tst2"
output: html_document
---
```{r setup, include=FALSE}
knitr::opts_chunk$set(echo = TRUE)
```
## R Markdown
This is an R Markdown document. Markdown is a simple formatting syntax for authoring HTML, PDF, and MS Word documents. For more details on using R Markdown see <http://rmarkdown.rstudio.com>.
When you click the **Knit** button a document will be generated that includes both content as well as the output of any embedded R code chunks within the document. You can embed an R code chunk like this:
```{r cars}
summary(cars)
```
## Including Plots
You can also embed plots, for example:
```{r pressure, echo=FALSE}
plot(pressure)
```
Note that the `echo = FALSE` parameter was added to the code chunk to prevent printing of the R code that generated the plot.
Session info
> sessionInfo()
R version 4.1.2 (2021-11-01)
Platform: x86_64-redhat-linux-gnu (64-bit)
Running under: Red Hat Enterprise Linux 8.5 (Ootpa)
Matrix products: default
BLAS/LAPACK: /usr/lib64/libopenblas-r0.3.12.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8
[4] LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
CodePudding user response:
I think the problem will be, that you are copying your prot.Rmd to a tempfile with fileext = ".html" - it should be ".Rmd".
Please try the following:
# Define UI for application that draws a histogram
ui <- fluidPage(
# Application title
titlePanel("Knit"),
downloadButton("prot", "Render file")
)
# Define server logic required to draw a histogram
server <- function(input, output) {
output$prot <- downloadHandler(
# For PDF output, change this to "report.pdf"
filename = "prot.html",
content = function(file) {
report_path <- tempfile(fileext = ".Rmd")
file.copy("prot.Rmd", report_path, overwrite = TRUE)
rmarkdown::render(report_path,
output_file = file)
}
)
}
# Run the application
shinyApp(ui = ui, server = server)
