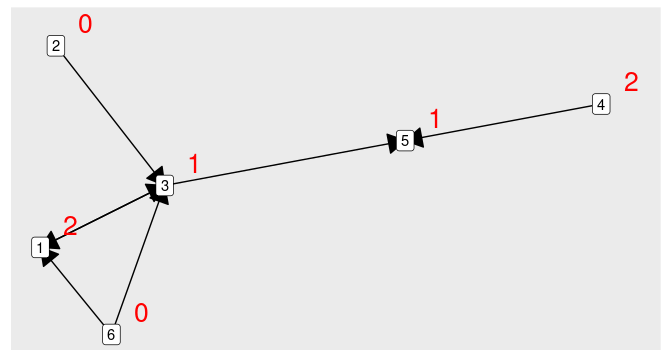
I have a graph with each node having a value (value in red).
I would like to do the following two things (I guess 1 is a special case of 2):
Each node should be assigned the mean of the value of the direct peers directing to it. For example node #5
(1 2)/2=1.5or node #3(0 2 0)/3=2/3.Instead of direct neighbors, include all connected nodes but with a diffusion of times 1/n with n being the distance to the node. The further away the information is coming from the weaker signal we'd have.
I looked into functions of igraph, but could not find anything that is doing this (I might have overseen though). How could I do this computation?
Below is the code for a sample network with random values.
library(tidyverse)
library(tidygraph)
library(ggraph)
set.seed(6)
q <- tidygraph::play_erdos_renyi(6, p = 0.2) %>%
mutate(id = row_number(),
value = sample(0:3, size = 6, replace = T))
q %>%
ggraph(layout = "with_fr")
geom_edge_link(arrow = arrow(length = unit(0.2, "inches"),
type = "closed"))
geom_node_label(aes(label = id))
geom_node_text(aes(label = value), color = "red", size = 7,
nudge_x = 0.2, nudge_y = 0.2)
Edit, found a solution to 1
q %>%
mutate(value_smooth = map_local_dbl(order = 1, mindist = 1, mode = "in",
.f = function(neighborhood, ...) {
mean(as_tibble(neighborhood, active = 'nodes')$value)
}))
CodePudding user response:
Each node should be assigned the mean of the value of the direct peers directing to it.
Guessing that you really mean
Each node should be assigned the mean of the values of the direct peers directing to it, before any node values were changed
This seems trivial - maybe I am missing something?
Loop over nodes
Sum values of adjacent nodes
Calculate mean and store in vector by node index
Loop over nodes
Set node value to mean stored in previous loop
CodePudding user response:
Probably you can try the code below
q %>%
set_vertex_attr(
name = "value",
value = sapply(
ego(., mode = "in", mindist = 1),
function(x) mean(x$value)
)
)
which gives
# A tbl_graph: 6 nodes and 7 edges
#
# A directed simple graph with 1 component
#
# Node Data: 6 x 2 (active)
id value
<int> <dbl>
1 1 0.5
2 2 NaN
3 3 0.667
4 4 NaN
5 5 1.5
6 6 NaN
#
# Edge Data: 7 x 2
from to
<int> <int>
1 3 1
2 6 1
3 1 3
# ... with 4 more rows