I want to create boxplots of to compare two groups' 5 continuous variable measurements named tics1, tics2, tics3, tics4, tics5. I can easily do that with this code:
boxplot(tics1 ~ group, data=tics, col=c("hotpink", "cyan2"))
boxplot(tics2 ~ group, data=tics, col=c("hotpink", "cyan2"))
boxplot(tics3 ~ group, data=tics, col=c("hotpink", "cyan2"))
boxplot(tics4 ~ group, data=tics, col=c("hotpink", "cyan2"))
boxplot(tics5 ~ group, data=tics, col=c("hotpink", "cyan2"))
But I'm trying to use a for loop to be more efficient. When I try this, I get an error.
for (i in 1:5) {
var <- paste0("tics", i)
boxplot(var ~ group, data=tics, col=c("hotpink", "cyan2"))
}
Error in stats::model.frame.default(formula = var ~ group, data = tics) : variable lengths differ (found for 'group')
- Is there a way to fix my for loop code?
- Is there a way to have all 5 comparisons on one boxplot?
CodePudding user response:
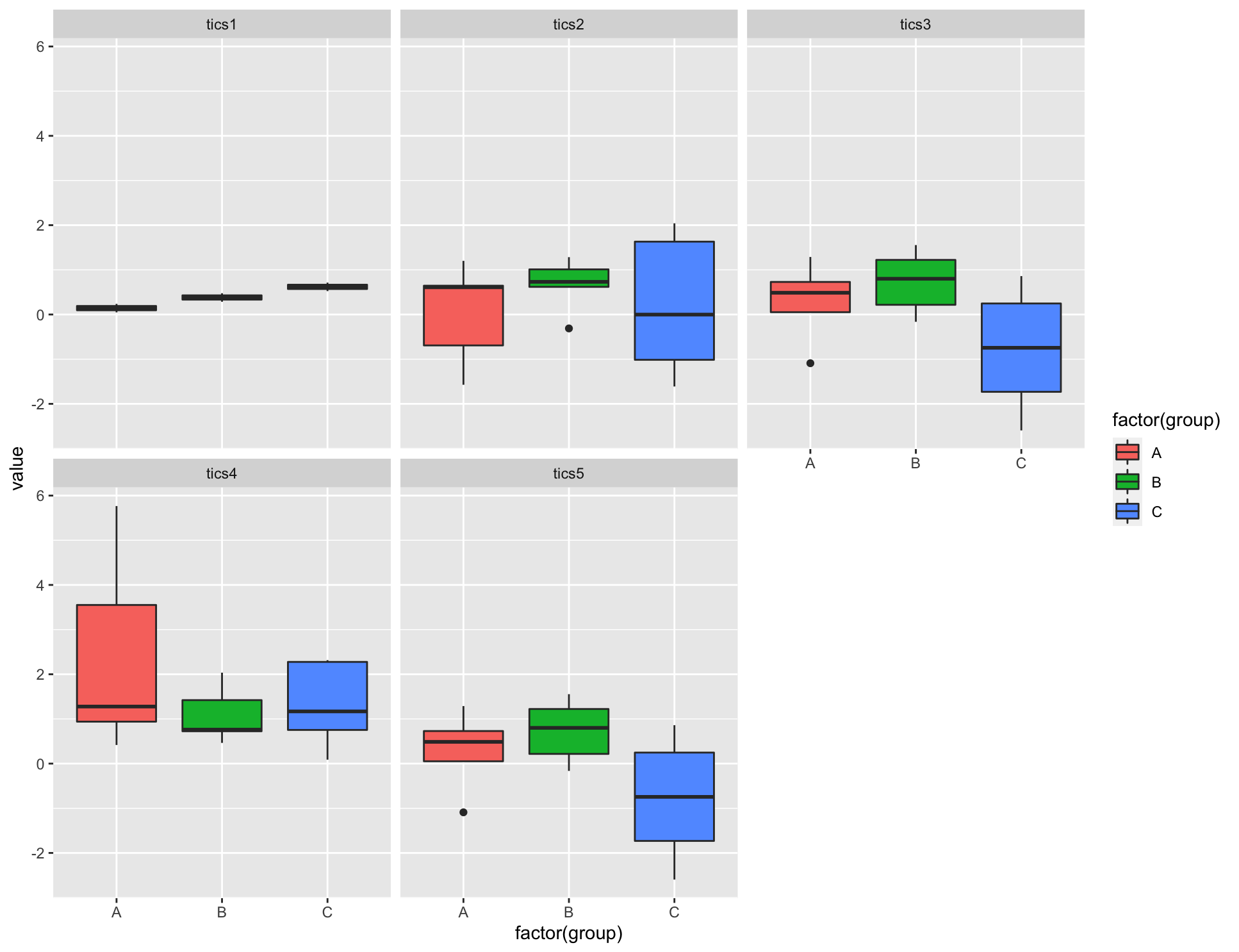
If you want to plot them all, then you can use facet_wrap from ggplot2. You would want to pivot to long format, then you can plot.
library(tidyverse)
tics %>%
pivot_longer(-group) %>%
ggplot(aes(x = factor(group), y = value, fill = factor(group)))
geom_boxplot()
facet_wrap(~name)
Output
Or with your for loop, you can do:
for (i in 1:5) {
var <- paste0("tics", i)
boxplot(tics[[var]] ~ group, data = tics, col=c("hotpink", "cyan2"))
}
If you go this route, then sapply would be quicker, and here I add the name to each plot as well.
sapply(1:5, \(x) {var <- paste0("tics", x); boxplot(tics[[var]] ~ tics$group, main = var)})
You could also loop by index, assuming that group is the first column and you only have tic columns in the dataframe.
for (i in 2:5) {
boxplot(mat[, i] ~ group, tics)
}
Data
tics <- structure(list(tics1 = c(0.0476190476190476, 0.0952380952380952,
0.142857142857143, 0.19047619047619, 0.238095238095238, 0.285714285714286,
0.333333333333333, 0.380952380952381, 0.428571428571429, 0.476190476190476,
0.523809523809524, 0.571428571428571, 0.619047619047619, 0.666666666666667,
0.714285714285714), tics2 = c(-0.692143884081275, 0.644709708117294,
-1.57303517336961, 1.20119221027555, 0.609239967840388, -0.311524439591859,
0.618602249192469, 0.731306188818431, 1.01016469827886, 1.28385223013644,
-0.00178540309357942, 2.041746200149, -1.01431257489833, -1.61190976820524,
1.63099766889229), tics3 = c(0.0520219824975517, 0.729165269851886,
1.28805775316925, -1.09043323687797, 0.486936194669402, 0.800131923610429,
1.22229153795252, 0.217159233531646, -0.163640790378808, 1.55459728200125,
0.860175585334737, -1.73107965801683, -0.744770481693222, -2.59518985923938,
0.246772490830949), tics4 = c(1.27763384585271, 0.939207828308425,
5.76608257808322, 0.416700865464712, 3.55156271227215, 0.463652374864707,
1.42103094782663, 0.724411125077308, 2.03621888478233, 0.760893978643801,
0.75365623199256, 2.31626695810966, 0.0881069629466973, 1.16878624674157,
2.27680839967629), group = c("A", "A", "A", "A", "A", "B", "B",
"B", "B", "B", "C", "C", "C", "C", "C"), tics5 = c(0.0520219824975517,
0.729165269851886, 1.28805775316925, -1.09043323687797, 0.486936194669402,
0.800131923610429, 1.22229153795252, 0.217159233531646, -0.163640790378808,
1.55459728200125, 0.860175585334737, -1.73107965801683, -0.744770481693222,
-2.59518985923938, 0.246772490830949)), row.names = c(NA, -15L
), class = "data.frame")