I did simulations in R and plotted the results in histograms. There is no problem adding probability density into the histograms with the density() function. But for some reason I get very strange results when I plot the theoretical densities to the histograms for comparison purposes. Here are two example codes and two pictures. The blue theoretical pdfs are way off and I do not know why. Could someone with R skills point out my mistakes?
# generating 10000 samples thrice from U(0,1) distribution
# and sorting them for the statistics:
n <- 10000
samples1 <- data.frame('0'=c(rep(NA,4)))
samples2 <- data.frame('0'=c(rep(NA,10)))
samples3 <- data.frame('0'=c(rep(NA,10)))
for (i in 1:n) {
new <- runif(4)
samples1[ , ncol(samples1) 1] <- sort(new)
colnames(samples1)[ncol(samples1)] <- i
new <- runif(10)
samples2[ , ncol(samples2) 1] <- sort(new)
colnames(samples2)[ncol(samples2)] <- i
new <- runif(10)
samples3[ , ncol(samples3) 1] <- sort(new)
colnames(samples3)[ncol(samples3)] <- i
}
# dropping the first (useless) columns:
samples1 <- samples1[-c(1)]
samples2 <- samples2[-c(1)]
samples3 <- samples3[-c(1)]
# selecting the statistics from the samples:
# X_2:4
stat24 <- rep(NA,n)
for (i in 1:n) {
stat24[i] <- samples1[2,i]
}
# X_2:10
stat210 <- rep(NA,n)
for (i in 1:n) {
stat210[i] <- samples1[2,i]
}
# X_10:10
stat1010 <- rep(NA,n)
for (i in 1:n) {
stat1010[i] <- samples1[10,i]
}
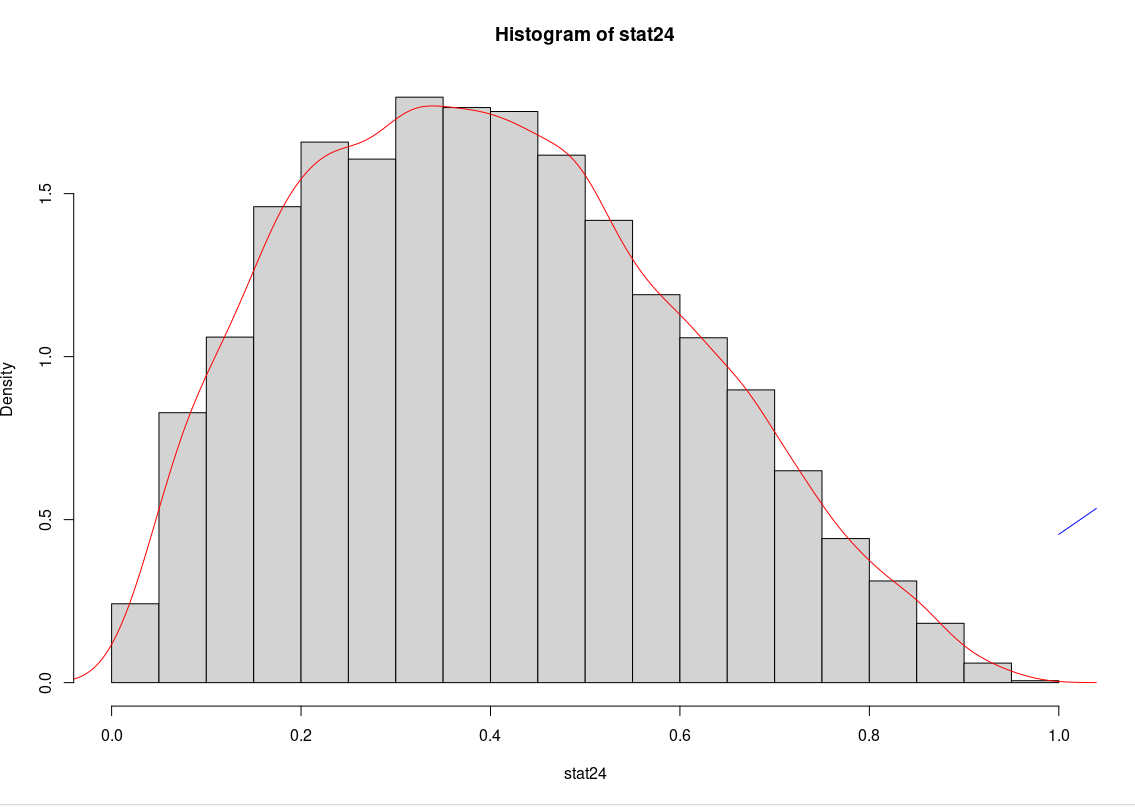
# plotting the histograms and Beta pdfs:
hist(stat24, freq = FALSE)
lines(dbeta(stat24, 2, 5), col='blue')
lines(density(stat24), col='red')
Distribution of the kth statistic follows Beta(k, n k-1) distribution which appears as the odd blue stroke on the right.
n <- 10000
random_variable_F <- rep(NA,n)
# generating 10000 samples of sizes 10 and 5 and computing F:
for (i in 1:n) {
x <- rnorm(10, mean = 10, sd = sqrt(5))
y <- rnorm(5, mean = 20, sd = sqrt(10))
random_variable_F[i] <- ((var(x))*5)/((var(y)*10))
}
#head(random_variable_F)
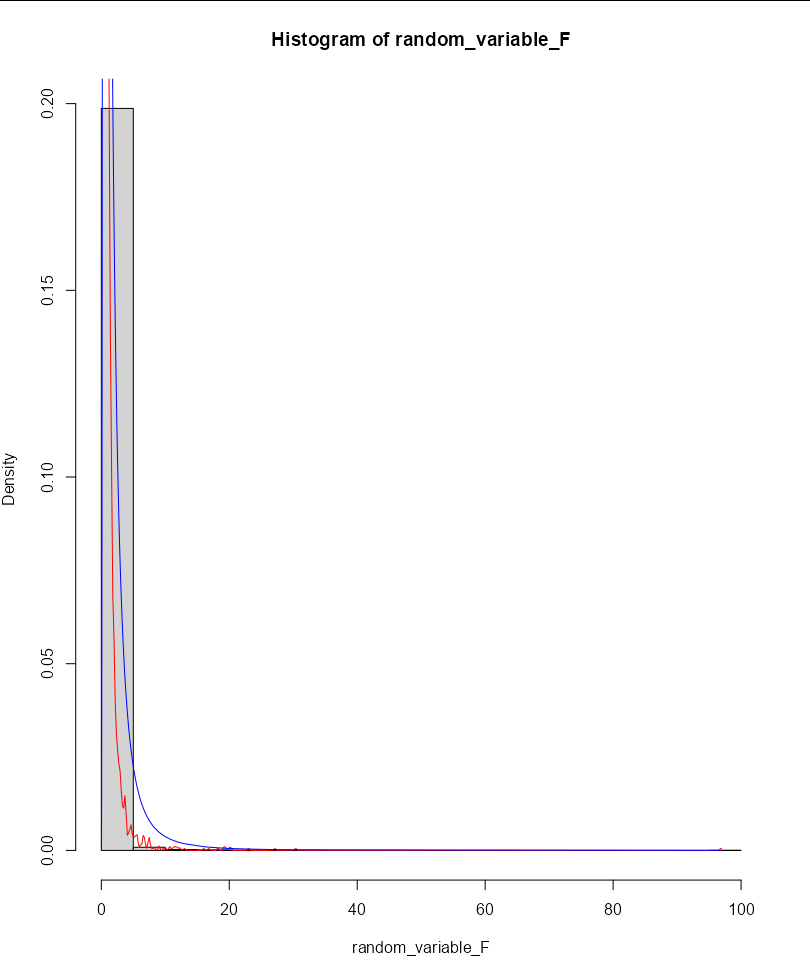
# plotting the histogram:
hist(random_variable_F, freq = F)
lines(density(random_variable_F), col='red')
lines(df(random_variable_F, 9, 4,), col='blue')
Random variable F follows F-distribution. Paramaters are the sample sizes minus one, in this case 10-1=9 and 5-1=4. the theoretical curve is quite wild:
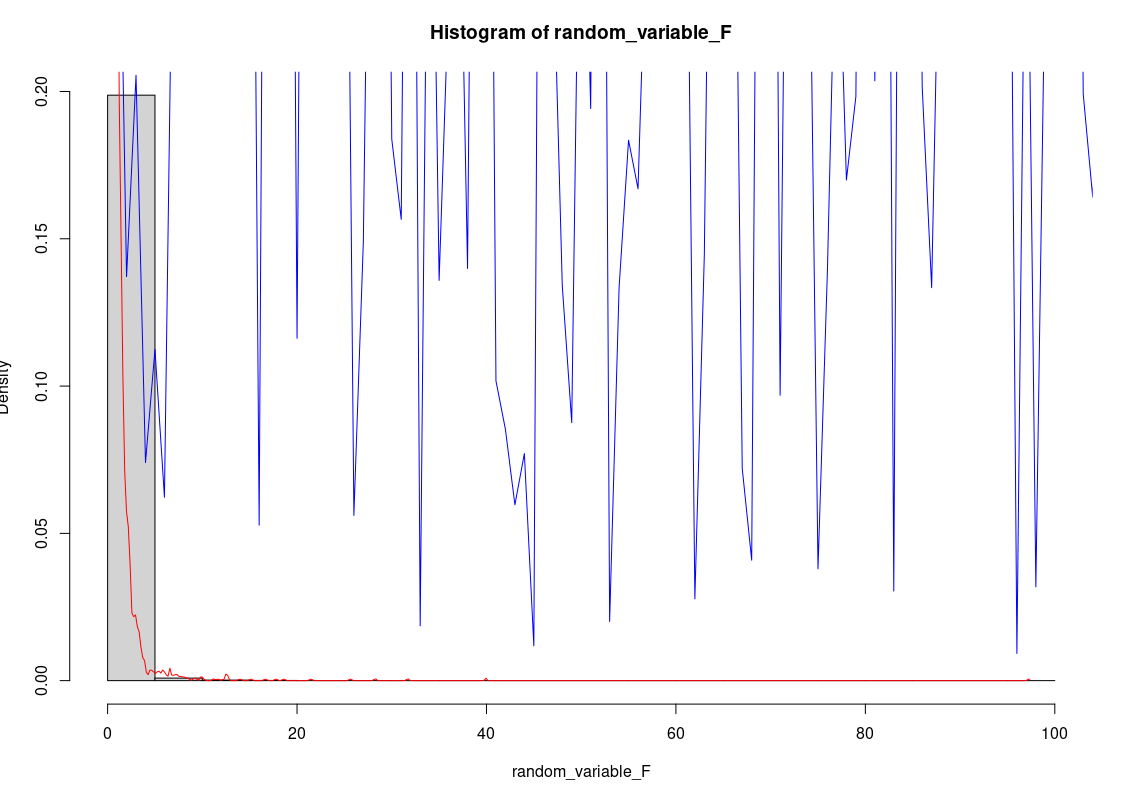
CodePudding user response:
If you pass a single vector to lines, it assumes that this is a vector of y values you want to plot. It plots the first y value at x = 1, the second y value at x = 2, etc, all the way up to x = length(y). In your case, random_variable_F is an unordered random variable, and you are just plotting its sequential values at 1:10000 along the x axis.
Clearly, you want the function y = df(x) to be plotted, so you need to pass random_variable_F as the x values and df(random_variable_F) as the y values. You will also need to sort random_variable_F first to ensure the line is plotted from left to right:
hist(random_variable_F, freq = F)
lines(density(random_variable_F), col='red')
lines(sort(random_variable_F), df(sort(random_variable_F), 9, 4), col='blue')
Note that this doesn't happen when you plot lines(density(random_variable_F)) because density produces a list containing ordered x and y valued rather than a vector.