I have two datasets.
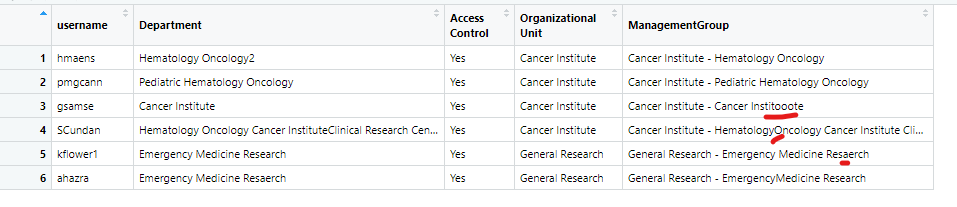
One is the data I'm currently working on and need to change (spelling mistakes), it looks like this:
df<-structure(list(username = c("hmaens", "pmgcann", "gsamse", "SCundan",
"kflower1", "ahazra"), Department = c("Hematology Oncology2",
"Pediatric Hematology Oncology", "Cancer Institute",
"Hematology Oncology Cancer InstituteClinical Research Center",
"Emergency Medicine Research", "Emergency Medicine Resaerch"),
`Access Control` = c("Yes", "Yes", "Yes", "Yes", "Yes", "Yes"
), `Organizational Unit` = structure(c(1L, 1L, 1L, 1L, 2L,
2L), .Label = c("Cancer Institute", "General Research"
), class = "factor"), ManagementGroup = c("Cancer Institute - Hematology Oncology",
"Cancer Institute - Pediatric Hematology Oncology",
"Cancer Institute - Cancer Institooote", "Cancer Institute - HematologyOncology Cancer Institute Clinical Research Center",
"General Research - Emergency Medicine Resaerch", "General Research - EmergencyMedicine Research"
)), row.names = c(NA, -6L), class = c("tbl_df", "tbl", "data.frame"
))
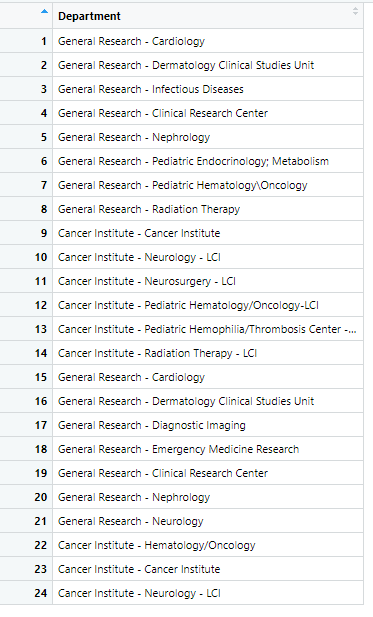
The other data frame is a reference list of how the Management groups should be spelled:
df2<-structure(list(Department = c("General Research - Cardiology ",
"General Research - Dermatology Clinical Studies Unit ", "General Research - Infectious Diseases ",
"General Research - Clinical Research Center ", "General Research - Nephrology ",
"General Research - Pediatric Endocrinology; Metabolism ", "General Research - Pediatric Hematology\\Oncology ",
"General Research - Radiation Therapy ", "Cancer Institute - Cancer Institute ",
"Cancer Institute - Neurology - LCI ", "Cancer Institute - Neurosurgery - LCI ",
"Cancer Institute - Pediatric Hematology/Oncology-LCI ",
"Cancer Institute - Pediatric Hemophilia/Thrombosis Center - LCI ",
"Cancer Institute - Radiation Therapy - LCI ", "General Research - Cardiology ",
"General Research - Dermatology Clinical Studies Unit ", "General Research - Diagnostic Imaging ",
"General Research - Emergency Medicine Research ", "General Research - Clinical Research Center ",
"General Research - Nephrology ", "General Research - Neurology ",
"Cancer Institute - Hematology/Oncology ", "Cancer Institute - Cancer Institute ",
"Cancer Institute - Neurology - LCI ")), row.names = c(NA,
-24L), class = c("tbl_df", "tbl", "data.frame"))
My question is: is there a way to automatically reference this second data frame for 'correct' spelling of words and change columns en masse?
I realize I could individually fix spelling mistakes with the methods used in this answer, where for instance I could write individual lines of code that say "no, resAErch should instead be spelled 'research'"... but is there a way to have R look for the 'nearest' spelling in that second data frame and change it to that?
Phrased another way, could R code be written that would check df$Managementgroup and notice that "Cancer Institute-Cancer Insitooote" is really similar to "Cancer Institute - Cancer Institute" found in df2$Department and then fix the spelling?
If that makes sense, ideally it would incorporate spelling and spaces from the second dataframe as well.
CodePudding user response:
this answer using {fuzzyjoin} is relevant. Good luck!
library(fuzzyjoin)
library(dplyr)
df <- structure(list(
username = c(
"hmaens", "pmgcann", "gsamse", "SCundan",
"kflower1", "ahazra"
),
Department = c(
"Hematology Oncology2",
"Pediatric Hematology Oncology", "Cancer Institute",
"Hematology Oncology Cancer InstituteClinical Research Center",
"Emergency Medicine Research", "Emergency Medicine Resaerch"
),
`Access Control` = c("Yes", "Yes", "Yes", "Yes", "Yes", "Yes"),
`Organizational Unit` = structure(c(
1L, 1L, 1L, 1L, 2L,
2L
), .Label = c("Cancer Institute", "General Research"), class = "factor"),
ManagementGroup = c(
"Cancer Institute - Hematology Oncology",
"Cancer Institute - Pediatric Hematology Oncology",
"Cancer Institute - Cancer Institooote", "Cancer Institute - HematologyOncology Cancer Institute Clinical Research Center",
"General Research - Emergency Medicine Resaerch", "General Research - EmergencyMedicine Research"
)
), row.names = c(NA, -6L), class = c("tbl_df", "tbl", "data.frame"))
df2 <- structure(list(ManagementGroup = c(
"General Research - Cardiology ",
"General Research - Dermatology Clinical Studies Unit ", "General Research - Infectious Diseases ",
"General Research - Clinical Research Center ", "General Research - Nephrology ",
"General Research - Pediatric Endocrinology; Metabolism ", "General Research - Pediatric Hematology\\Oncology ",
"General Research - Radiation Therapy ", "Cancer Institute - Cancer Institute ",
"Cancer Institute - Neurology - LCI ", "Cancer Institute - Neurosurgery - LCI ",
"Cancer Institute - Pediatric Hematology/Oncology-LCI ",
"Cancer Institute - Pediatric Hemophilia/Thrombosis Center - LCI ",
"Cancer Institute - Radiation Therapy - LCI ", "General Research - Cardiology ",
"General Research - Dermatology Clinical Studies Unit ", "General Research - Diagnostic Imaging ",
"General Research - Emergency Medicine Research ", "General Research - Clinical Research Center ",
"General Research - Nephrology ", "General Research - Neurology ",
"Cancer Institute - Hematology/Oncology ", "Cancer Institute - Cancer Institute ",
"Cancer Institute - Neurology - LCI "
)), row.names = c(
NA,
-24L
), class = c("tbl_df", "tbl", "data.frame"))
final_df <- stringdist_join(df, df2,
by = "ManagementGroup",
mode = "left",
ignore_case = FALSE,
method = "jw",
max_dist = 99,
distance_col = "dist") %>%
group_by(ManagementGroup.x) %>%
slice_min(order_by = dist, n = 1) %>%
distinct()
Created on 2022-04-05 by the reprex package (v2.0.1)