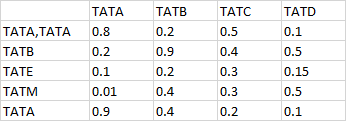
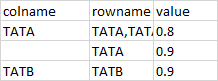
I have a vector with 3990 names (these are the column names of my dataframe) and I want to match them with the rows of my data. My data contains correlation values and I want to subset my data based on the matches found
I tried using grepl
result <- filter(df, grepl(paste(column_names, collapse="|"), rownames(df)))
but I get an error
error in 'grepl()': ! invalid regular expression
my expected output would be
does anyone have any suggestions on how can this be done?
would really be great if someone could help me with this!
Best, Shweta
CodePudding user response:
Try this:
library(dplyr)
dat %>%
filter(grepl(paste0("\\b", names(.), "\\b", collapse="|"), rownames(dat)))
TATA TATB TATC TATD
TATA,TATA 0.8 0.2 0.5 0.1
TATB 0.2 0.9 0.4 0.5
TATA 0.9 0.4 0.2 0.1
Data:
dat <- data.frame(TATA = c(0.8,0.2,0.1,0.01,0.9),
TATB = c(0.2,0.9,0.2,0.4,0.4),
TATC = c(0.5,0.4,0.3,0.3,0.2),
TATD = c(0.1,0.5,0.15,0.5,0.1),
row.names = c("TATA,TATA", "TATB", "TATE", "TATM", "TATA"))
CodePudding user response:
Three options you can try:
library(reshape2)
melt(as.matrix(dat)
data.frame(rows=rownames(dat)[row(dat)], vars=colnames(dat)[col(dat)], values=c(dat))
as.data.frame(as.table(as.matrix(dat)))
result:
Var1 Var2 Freq
1 TATA,TATA TATA 0.872624483
2 TATB TATA 0.533790730
3 TATE TATA 0.110495616
4 TATM TATA 0.253893718
5 TATA TATA 0.303576730
6 TATA,TATA TATB 0.774815753
7 TATB TATB 0.941361633
8 TATE TATB 0.305219935
9 TATM TATB 0.101124692
10 TATA TATB 0.968514156
11 TATA,TATA TATC 0.891697937
12 TATB TATC 0.006223573
13 TATE TATC 0.045138657
14 TATM TATC 0.848485971
15 TATA TATC 0.995542845
16 TATA,TATA TATD 0.479559761
17 TATB TATD 0.981808763
18 TATE TATD 0.227518091
19 TATM TATD 0.767491049
20 TATA TATD 0.410935185
data:
dat <- data.frame(TATA = runif(5),
TATB = runif(5),
TATC = runif(5),
TATD = runif(5),
row.names = c("TATA,TATA", "TATB", "TATE", "TATM", "TATA"))
EDIT:
Using regex matching to subset your input data, as a first step:
cols <- grep(pattern = paste0(rownames(dat), collapse = "|"), x = colnames(dat), value = TRUE)
rows <- grep(pattern = paste0(colnames(dat), collapse = "|"), x = rownames(dat), value = TRUE)
dat2 <- dat[rownames(dat) %in% rows, colnames(dat) %in% cols]
Yielding:
as.data.frame(as.table(as.matrix(dat2)))
Var1 Var2 Freq
1 TATA,TATA TATA 0.6908219
2 TATB TATA 0.7255142
3 TATA TATA 0.1022963
4 TATA,TATA TATB 0.7291625
5 TATB TATB 0.7420069
6 TATA TATB 0.7480157