I have a list of character vectors that I would like to convert into a tidy data frame. The lengths of the character vectors are unequal.
dput(data)
list(`ko03008 Ribosome biogenesis in eukaryotes` = c("G5382",
"G13330", "G4043", "G13255"), `ko03010 Ribosome` = c("G16823",
"G4822", "G11737", "G114", "G18144", "G6031", "G24182", "G9882",
"G14270", "G16903", "G2506", "G3550"), `ko03013 RNA transport` = c("G18058",
"G20817", "G6913", "G18004", "G4129", "G5382", "G5264", "G17529",
"G5114", "G21371", "G19351", "G15511", "G1049", "G14663"), `ko03015 mRNA surveillance pathway` = c("G20817",
"G6913", "G18004", "G4129", "G5382", "G19351", "G15511", "G1463"
), `ko03018 RNA degradation` = c("G11453", "G7437", "G11483",
"G12095"), `ko03020 RNA polymerase` = c("G13069", "G10917", "G6973",
"G7432"))
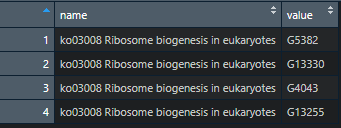
I would like to create a data frame with two columns. One with the name of each character vector within the list (e.g. 'ko03008 Ribosome biogeneis in eukaryotes') and the other with gene IDs (e.g. 'G5382).
I've used enframe to create a tibble that looks like this:
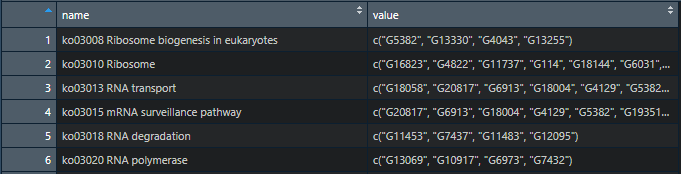
but I would like to format it like this (an example of what the first vector in the list should look like):
CodePudding user response:
Use unnest_longer:
library(tidyverse)
data %>%
enframe() %>%
unnest_longer(value)
# A tibble: 46 x 2
name value
<chr> <chr>
1 ko03008 Ribosome biogenesis in eukaryotes G5382
2 ko03008 Ribosome biogenesis in eukaryotes G13330
3 ko03008 Ribosome biogenesis in eukaryotes G4043
4 ko03008 Ribosome biogenesis in eukaryotes G13255
5 ko03010 Ribosome G16823
6 ko03010 Ribosome G4822
7 ko03010 Ribosome G11737
8 ko03010 Ribosome G114
9 ko03010 Ribosome G18144
10 ko03010 Ribosome G6031
# ... with 36 more rows