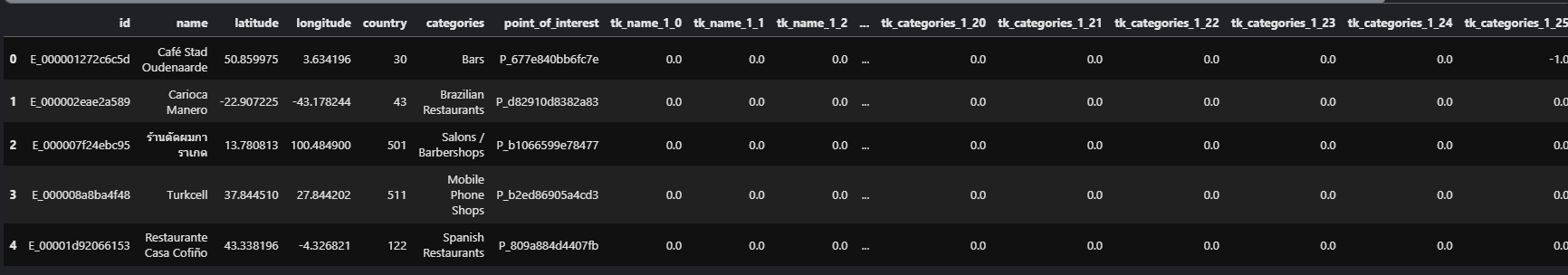
I have a DataFrame test with shape (1138812, 57). The head looks like this:
And I have an array indices which has a shape (1138812, 25). It is a 2D array with each subarray having 25 indices. It looks like this:
[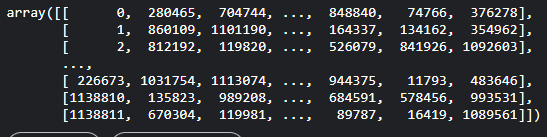
the indices array has 25 indices from the DataFrame corresponding to each 1138812 indices from the same DataFrame. I want to create a new DataFrame with 25 X 1138812 rows based on this array. For example:
i have a 2d array, something like:
[[0,2,3],
[1,0,3],
[2,1,0],
[3,1,2]]
and i have a pandas dataframe something like:
id val
0 a 9
1 b 8
2 c 3
3 d 7
now i want to get a new dataframe based on the indexes listed in the 2d array, for this example, it will be like:
id val id_2 val
0 a 9 a 9
0 a 9 c 3
0 a 9 d 7
1 b 8 b 8
1 b 8 a 9
1 b 9 d 7
2 c 3 c 3
2 c 3 b 8
2 c 3 a 9
3 d 7 d 7
3 d 7 b 8
3 d 7 c 3
I tried many approaches including:
import pandas as pd
import numpy as np
index = [[0,2,3],
[1,0,3],
[2,1,0],
[3,1,2]]
idse = ['a','b','c','d']
vals = [9,8,3,7]
data = {'id': idse, 'val': vals}
df = pd.DataFrame(data=data)
newdf = pd.DataFrame(np.repeat(df.values, len(index[0]), axis=0))
flat_list = [item for sublist in index for item in sublist]
newdf['id_2'] = df.id[flat_list].values
newdf['val_2'] = df.val[flat_list].values
and
fdf = pd.DataFrame()
for i, ir in enumerate(l):
temp_df = df.iloc[ir]
temp_df['id'] = df.iloc[i]['id']
temp_df = pd.merge(df,temp_df,how="outer",on="id")
temp_df = temp_df[temp_df['id']==df.iloc[i]['id']]
fdf = pd.concat([fdf,temp_df])
fdf
both of them work the way I want but they are very very slow for the original DataFrame with 1.1m rows and they take up all the ram which crashes the notebook. I am using RAPIDS libraries including cuDF, cuPy, cuML which correspond to pandas, numpy/scipy and sklearn respectively and I need a pure numpy/pandas solution so that they can use the GPU cores and make this operation quicker and more efficient.
Thanks
CodePudding user response:
Let us try assign with explode then join
out = df.assign(new=a.tolist()).explode('new').set_index('new').add_suffix('_2').join(df)
CodePudding user response:
Assuming df and a the input dataframe and array, you can repeat the indices of your dataframe and concat it with the dataframes indexed from the flattened array:
idx = df.index.repeat(a.shape[1])
df2 = pd.concat(
[df.loc[idx],
df.loc[a.ravel()].add_suffix('_2').set_axis(idx)
], axis=1)
output:
id val id_2 val_2
0 a 9 a 9
0 a 9 c 3
0 a 9 d 7
1 b 8 b 8
1 b 8 a 9
1 b 8 d 7
2 c 3 c 3
2 c 3 b 8
2 c 3 a 9
3 d 7 d 7
3 d 7 b 8
3 d 7 c 3
used input:
df = pd.DataFrame({'id': ['a', 'b', 'c', 'd'],
'val': [9, 8, 3, 7]})
a = np.array([[0,2,3],
[1,0,3],
[2,1,0],
[3,1,2]])
NB. a quick test shows that is takes 900ms to process 1M rows