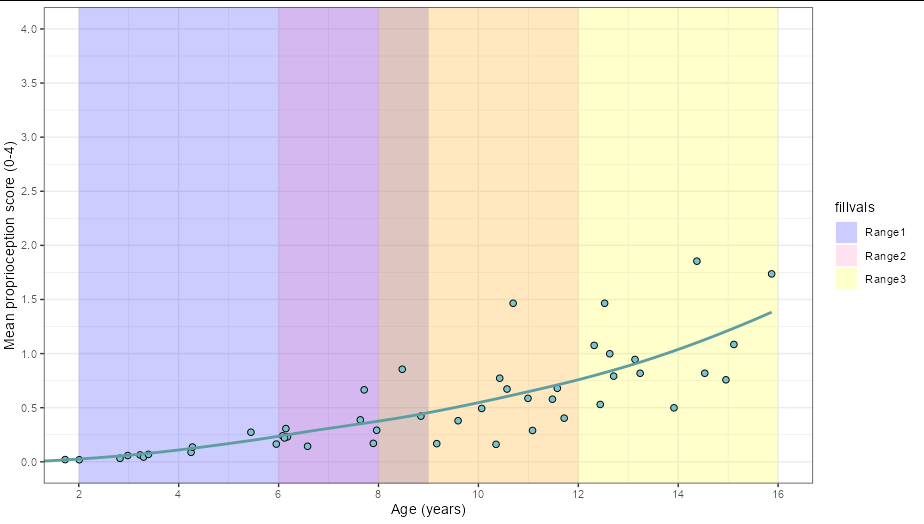
I have created a scatterplot using ggplot2 in RStudio with the function annotate("rect".... to add a layer of colours based on another variable. I have been trying to add a legend for SO long by googling and trying different methods found on this site and others, including using geom_rect function but I cannot seem to get it to work fully. Please can anyone help me add a legend purely based on the shaded areas. See my plot:
This is my script so far:
ggplot(prpc_data,aes(x=age,y=total_proprioception))
annotate("rect",xmin = 2, xmax = 9, ymin = -Inf, ymax = Inf, colour="blue", fill="blue",alpha = 0.15)
annotate("rect",xmin=6,xmax=12,ymin=-Inf,ymax=Inf,colour="hotpink",fill="hotpink",alpha=0.15)
annotate("rect",xmin=8,xmax=16,ymin=-Inf,ymax=Inf,colour="yellow",fill="yellow",alpha=0.2)
geom_point(shape=21,size=2,colour="black",fill="cadetblue3")
geom_smooth(method="loess",se=FALSE,colour="cadetblue")
labs(x="Age (years)",y="Mean proprioception score (0-4)")
coord_cartesian(ylim=c(0,4),xlim=c(2,16))
scale_y_continuous(breaks=seq(0,4,0.5))
scale_x_continuous(breaks=seq(2,16,2))
theme_bw()
I would be very grateful for any help
CodePudding user response:
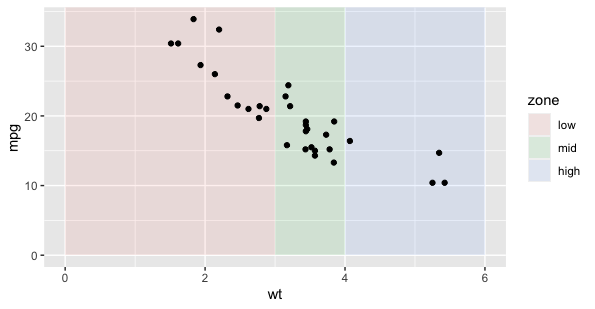
Annotation layers in ggplot2 are typically used for one-off labels, notes, and symbols, and don't usually relate to a legend. The easiest way to get a legend for some shaded areas would be to make those a normal geom_* layer with aethestics mapped to variables.
Below, I give the geom_rect layer its own data frame, and specific inherit.aes = FALSE so that that layer ignores the global aes(wt, mpg) assignment. Since the fill of those rectangles are mapped to the category, a legend is automatically created.
ggplot(mtcars, aes(wt, mpg))
geom_rect(data = data.frame(zone = factor(c("low", "mid", "high"),
levels = c("low", "mid", "high")),
start = c(0,3,4),
end = c(3,4,6)),
inherit.aes = FALSE,
aes(xmin = start, xmax = end, ymin = 0, ymax = Inf, fill = zone),
alpha = 0.1)
geom_point()
CodePudding user response:
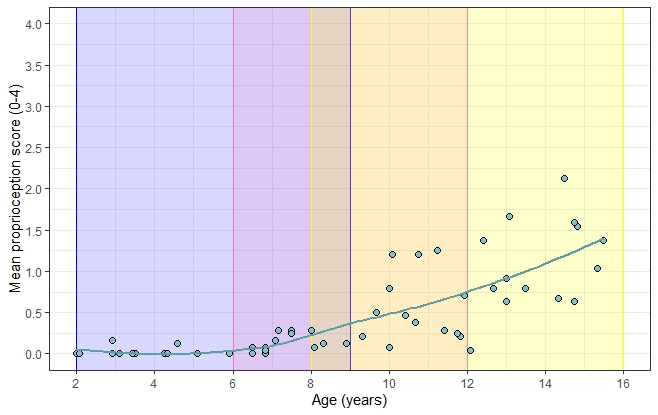
Instead of adding rectangles using annotate, you can use geom_rect and map its fill aesthetic to whatever labels you want. To do this you can create a little rect_df that you pass to geom_rect, and use scale_fill_manual to give the rectangles the fill colours you want:
rect_df <- data.frame(xmin = c(2, 6, 8), xmax = c(9, 12, 16),
labs = c("Range1", "Range2", "Range3"))
ggplot(prpc_data, aes(age, total_proprioception))
geom_rect(aes(xmin = xmin, xmax = xmax, ymin = -Inf, ymax = Inf, fill = labs),
alpha = 0.2, data = rect_df, inherit.aes = FALSE)
geom_point(shape = 21, size = 2, colour = "black", fill = "cadetblue3")
geom_smooth(method = "loess", se = FALSE, colour = "cadetblue")
labs(x = "Age (years)", y = "Mean proprioception score (0-4)")
coord_cartesian(ylim = c(0, 4), xlim = c(2, 16))
scale_fill_manual(values = c("blue", "hotpink", "yellow"))
scale_y_continuous(breaks = seq(0, 4, 0.5))
scale_x_continuous(breaks = seq(2, 16, 2))
theme_bw()
Data used to emulate OP plot
set.seed(1)
prpc_data <- data.frame(age = runif(50, 0, 16))
prpc_data$total_proprioception <- rnorm(50, prpc_data$age, prpc_data$age/4)^2/200