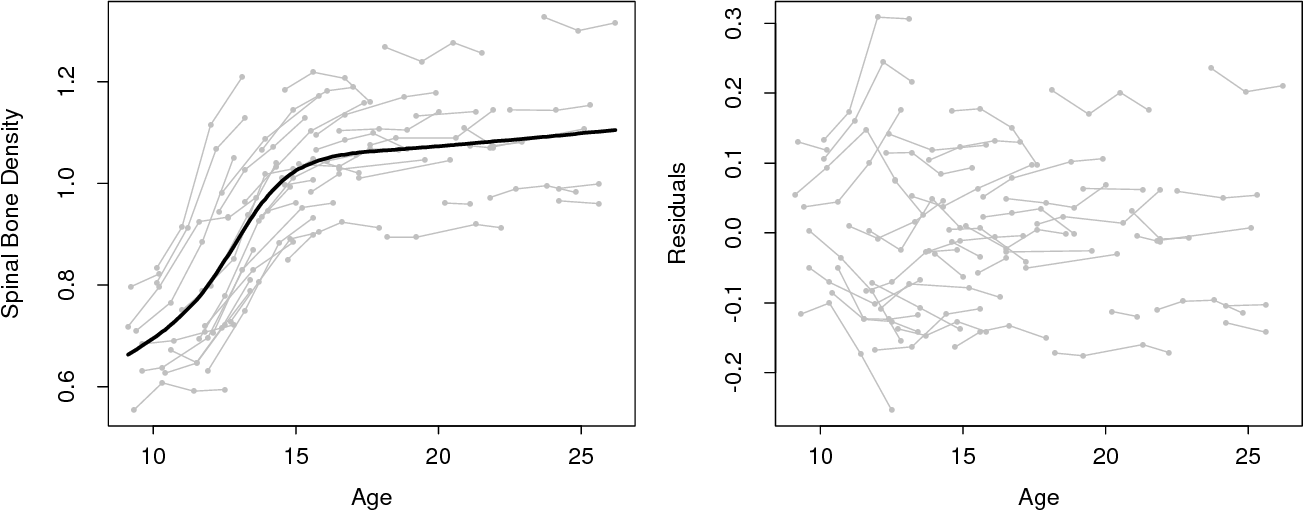
I'm wondering how to reproduce the following figure using R.
The data used in the figure are sparse functional data of bone mineral density. Basically each participant's bone mineral level is observed a few times during the experiment. But the observation times and number of observations for each participant are different.
The figure is from article 'Principal component models for sparse functional data'.
You can find it here 
CodePudding user response:
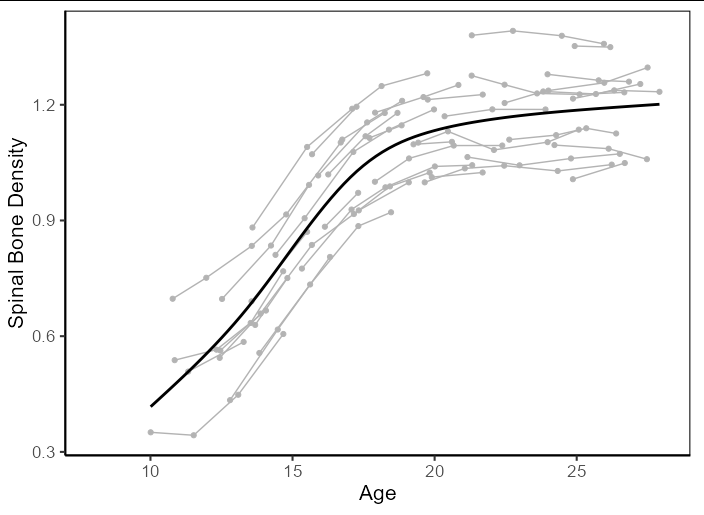
You could reproduce the figure with made-up data like this:
library(ggplot2)
# Create sample data
set.seed(8) # Makes data reproducible
ages <- runif(40, 8, 24)
df <- do.call(rbind, lapply(seq_along(ages), function(x) {
age <- ages[x] cumsum(runif(sample(2:5, 1), 1, 2))
y <- (tanh((age - 10)/pi - pi/2) 2.5)/3
y <- y rnorm(1, 0, 0.1)
y <- y cumsum(rnorm(length(y), 0, 0.02))
data.frame(ID = x, age = age, BMD = y)
}))
# Draw plot
ggplot(df, aes(x = age, y = BMD))
geom_path(aes(group = ID), color = 'gray70', na.rm = TRUE)
geom_point(color = 'gray70', na.rm = TRUE)
geom_smooth(color = 'black', se = FALSE, formula =y ~ s(x, bs = "cs"),
method = 'gam', na.rm = TRUE)
theme_classic(base_size = 16)
scale_x_continuous(limits = c(8, 28))
labs(y = 'Spinal Bone Density', x = 'Age')
theme(panel.border = element_rect(fill = NA))
Without knowing your own data structure however, it's difficult to say how applicable you will find this to your own use case.